
Ranid herpesvirus 3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Alloherpesviridae; Batrachovirus
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
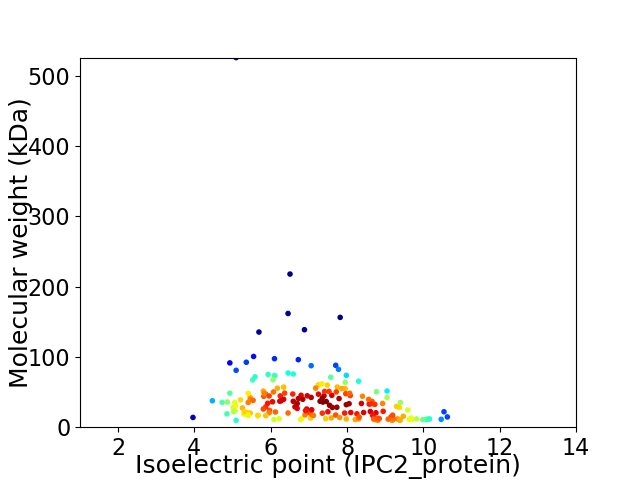
Virtual 2D-PAGE plot for 186 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X9T5L2|A0A1X9T5L2_9VIRU C-type lectin protein OS=Ranid herpesvirus 3 OX=1987509 PE=4 SV=1
MM1 pKa = 7.39SFAVDD6 pKa = 3.51DD7 pKa = 4.25RR8 pKa = 11.84LVSVVVIVGTAVFTVVMVTVGIGAFVVVVTICVAVALVVTLAFVVAIIVAVALFVTLAFVVAIIVAVEE76 pKa = 3.74LVITLAFVVAIVALMVVLIAEE97 pKa = 4.68AVTSVSSVLVDD108 pKa = 3.56GTTVEE113 pKa = 4.26GFVMGKK119 pKa = 10.25GDD121 pKa = 3.79PTSVQNDD128 pKa = 3.34EE129 pKa = 4.16PAFMISKK136 pKa = 10.29
MM1 pKa = 7.39SFAVDD6 pKa = 3.51DD7 pKa = 4.25RR8 pKa = 11.84LVSVVVIVGTAVFTVVMVTVGIGAFVVVVTICVAVALVVTLAFVVAIIVAVALFVTLAFVVAIIVAVEE76 pKa = 3.74LVITLAFVVAIVALMVVLIAEE97 pKa = 4.68AVTSVSSVLVDD108 pKa = 3.56GTTVEE113 pKa = 4.26GFVMGKK119 pKa = 10.25GDD121 pKa = 3.79PTSVQNDD128 pKa = 3.34EE129 pKa = 4.16PAFMISKK136 pKa = 10.29
Molecular weight: 13.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X9T569|A0A1X9T569_9VIRU Uncharacterized protein OS=Ranid herpesvirus 3 OX=1987509 PE=4 SV=1
MM1 pKa = 7.21VSKK4 pKa = 10.69HH5 pKa = 5.59PPAYY9 pKa = 9.66SSGFFEE15 pKa = 4.24HH16 pKa = 7.45RR17 pKa = 11.84AMQSSLHH24 pKa = 5.04EE25 pKa = 4.3AKK27 pKa = 10.8GNVGSRR33 pKa = 11.84PATPPIAFSPRR44 pKa = 11.84VGAPHH49 pKa = 6.09VPSRR53 pKa = 11.84DD54 pKa = 3.04GFNQNTHH61 pKa = 6.65ALGQTQSGFSRR72 pKa = 11.84QNPGRR77 pKa = 11.84QPFKK81 pKa = 11.11SKK83 pKa = 10.28ISRR86 pKa = 11.84LQPIYY91 pKa = 9.08EE92 pKa = 4.29TQTLHH97 pKa = 7.48RR98 pKa = 11.84FVQRR102 pKa = 11.84EE103 pKa = 3.58ARR105 pKa = 11.84RR106 pKa = 11.84TIEE109 pKa = 4.65HH110 pKa = 5.95VGAPNEE116 pKa = 4.04PVPPGTFVSVSPTARR131 pKa = 11.84TGQNHH136 pKa = 4.98SRR138 pKa = 11.84EE139 pKa = 4.07NQRR142 pKa = 11.84NIITTTAGHH151 pKa = 6.73RR152 pKa = 11.84NLPTHH157 pKa = 5.58FVEE160 pKa = 5.11YY161 pKa = 10.83SRR163 pKa = 11.84ANPKK167 pKa = 9.75NLPDD171 pKa = 5.18LIPVEE176 pKa = 4.95RR177 pKa = 11.84YY178 pKa = 7.4EE179 pKa = 4.34FPKK182 pKa = 10.22SCRR185 pKa = 11.84GMDD188 pKa = 3.29SAGRR192 pKa = 11.84CQRR195 pKa = 11.84LFSGSFRR202 pKa = 11.84IFTGDD207 pKa = 3.22LYY209 pKa = 11.39EE210 pKa = 4.87DD211 pKa = 4.02GKK213 pKa = 11.08RR214 pKa = 11.84EE215 pKa = 4.02NIVII219 pKa = 4.11
MM1 pKa = 7.21VSKK4 pKa = 10.69HH5 pKa = 5.59PPAYY9 pKa = 9.66SSGFFEE15 pKa = 4.24HH16 pKa = 7.45RR17 pKa = 11.84AMQSSLHH24 pKa = 5.04EE25 pKa = 4.3AKK27 pKa = 10.8GNVGSRR33 pKa = 11.84PATPPIAFSPRR44 pKa = 11.84VGAPHH49 pKa = 6.09VPSRR53 pKa = 11.84DD54 pKa = 3.04GFNQNTHH61 pKa = 6.65ALGQTQSGFSRR72 pKa = 11.84QNPGRR77 pKa = 11.84QPFKK81 pKa = 11.11SKK83 pKa = 10.28ISRR86 pKa = 11.84LQPIYY91 pKa = 9.08EE92 pKa = 4.29TQTLHH97 pKa = 7.48RR98 pKa = 11.84FVQRR102 pKa = 11.84EE103 pKa = 3.58ARR105 pKa = 11.84RR106 pKa = 11.84TIEE109 pKa = 4.65HH110 pKa = 5.95VGAPNEE116 pKa = 4.04PVPPGTFVSVSPTARR131 pKa = 11.84TGQNHH136 pKa = 4.98SRR138 pKa = 11.84EE139 pKa = 4.07NQRR142 pKa = 11.84NIITTTAGHH151 pKa = 6.73RR152 pKa = 11.84NLPTHH157 pKa = 5.58FVEE160 pKa = 5.11YY161 pKa = 10.83SRR163 pKa = 11.84ANPKK167 pKa = 9.75NLPDD171 pKa = 5.18LIPVEE176 pKa = 4.95RR177 pKa = 11.84YY178 pKa = 7.4EE179 pKa = 4.34FPKK182 pKa = 10.22SCRR185 pKa = 11.84GMDD188 pKa = 3.29SAGRR192 pKa = 11.84CQRR195 pKa = 11.84LFSGSFRR202 pKa = 11.84IFTGDD207 pKa = 3.22LYY209 pKa = 11.39EE210 pKa = 4.87DD211 pKa = 4.02GKK213 pKa = 11.08RR214 pKa = 11.84EE215 pKa = 4.02NIVII219 pKa = 4.11
Molecular weight: 24.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
67043 |
89 |
4633 |
360.4 |
40.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.311 ± 0.116 | 2.822 ± 0.147 |
4.8 ± 0.154 | 5.431 ± 0.221 |
4.627 ± 0.146 | 4.596 ± 0.129 |
2.543 ± 0.096 | 5.869 ± 0.089 |
5.743 ± 0.153 | 9.856 ± 0.138 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.506 ± 0.077 | 5.565 ± 0.138 |
4.785 ± 0.155 | 3.459 ± 0.126 |
4.992 ± 0.136 | 7.54 ± 0.147 |
6.663 ± 0.179 | 6.409 ± 0.119 |
1.247 ± 0.087 | 4.236 ± 0.108 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
