
Desulfurivibrio alkaliphilus (strain DSM 19089 / UNIQEM U267 / AHT2)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobulbaceae; Desulfurivibrio; Desulfurivibrio alkaliphilus
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
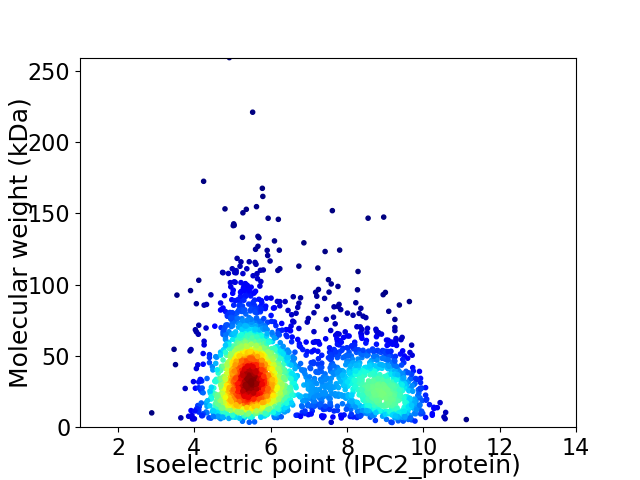
Virtual 2D-PAGE plot for 2595 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D6Z4S8|D6Z4S8_DESAT Beta-lactamase domain-containing protein OS=Desulfurivibrio alkaliphilus (strain DSM 19089 / UNIQEM U267 / AHT2) OX=589865 GN=DaAHT2_1873 PE=4 SV=1
MM1 pKa = 7.18SQRR4 pKa = 11.84PLLQPVVFLAALVCLLAVTFWSAPPATAAEE34 pKa = 4.32EE35 pKa = 4.36VATVVALRR43 pKa = 11.84GTAVAVDD50 pKa = 3.19AAGAEE55 pKa = 3.82RR56 pKa = 11.84ALAIKK61 pKa = 10.48APIHH65 pKa = 5.65RR66 pKa = 11.84QDD68 pKa = 3.83TIRR71 pKa = 11.84TGPRR75 pKa = 11.84GRR77 pKa = 11.84LQLMFTDD84 pKa = 3.63NTIISLGVNAEE95 pKa = 4.16LEE97 pKa = 4.02IAEE100 pKa = 4.4YY101 pKa = 10.56QWDD104 pKa = 3.86QQEE107 pKa = 4.82GEE109 pKa = 4.22LNTRR113 pKa = 11.84VNQGAFRR120 pKa = 11.84VMGGSVARR128 pKa = 11.84TSPEE132 pKa = 3.64KK133 pKa = 10.28FTTDD137 pKa = 2.92TPAATIGIRR146 pKa = 11.84GSMYY150 pKa = 10.44AGRR153 pKa = 11.84VSDD156 pKa = 3.68GALAVVFQGGTGIIVSNPAGTVEE179 pKa = 3.81ITTPGMGTRR188 pKa = 11.84VASRR192 pKa = 11.84TAAPTPPTVFTQEE205 pKa = 4.06DD206 pKa = 3.9LGALEE211 pKa = 5.0DD212 pKa = 4.06EE213 pKa = 4.84EE214 pKa = 4.74VDD216 pKa = 3.51EE217 pKa = 4.6EE218 pKa = 4.2AAEE221 pKa = 4.18EE222 pKa = 4.15EE223 pKa = 4.56TVEE226 pKa = 4.7DD227 pKa = 3.8EE228 pKa = 4.73TPAEE232 pKa = 4.18EE233 pKa = 4.16PAEE236 pKa = 4.24EE237 pKa = 4.19EE238 pKa = 4.34WEE240 pKa = 4.13DD241 pKa = 3.53AALDD245 pKa = 4.03EE246 pKa = 5.0PEE248 pKa = 4.09EE249 pKa = 4.11AAAPVAAAPAPITDD263 pKa = 3.8TAVDD267 pKa = 3.64QATEE271 pKa = 4.24TTTDD275 pKa = 3.23AAQDD279 pKa = 3.25EE280 pKa = 4.78WEE282 pKa = 4.28QTVVEE287 pKa = 4.81ADD289 pKa = 4.16PEE291 pKa = 4.31PTDD294 pKa = 4.65PDD296 pKa = 3.72PTDD299 pKa = 4.37PEE301 pKa = 5.56PIDD304 pKa = 4.6PEE306 pKa = 4.05PTDD309 pKa = 3.66PSEE312 pKa = 3.88AVYY315 pKa = 10.33SGRR318 pKa = 11.84GIEE321 pKa = 4.44IIYY324 pKa = 10.58SSMQEE329 pKa = 4.01EE330 pKa = 4.81VAVEE334 pKa = 3.93HH335 pKa = 6.82LVLTDD340 pKa = 3.82KK341 pKa = 10.9PSEE344 pKa = 4.02LTEE347 pKa = 4.14IEE349 pKa = 4.45AGLLEE354 pKa = 4.39YY355 pKa = 10.97NEE357 pKa = 4.32QSDD360 pKa = 4.17FSDD363 pKa = 3.92GEE365 pKa = 4.34YY366 pKa = 10.53SAVWSYY372 pKa = 11.82TDD374 pKa = 3.27ATQVEE379 pKa = 4.87EE380 pKa = 4.48WEE382 pKa = 4.51EE383 pKa = 4.27GDD385 pKa = 5.54DD386 pKa = 5.09FDD388 pKa = 5.32FQFTFVMVDD397 pKa = 3.21DD398 pKa = 5.35RR399 pKa = 11.84GEE401 pKa = 3.81FWLYY405 pKa = 8.31EE406 pKa = 3.92QWSDD410 pKa = 4.08TEE412 pKa = 4.37TDD414 pKa = 3.56EE415 pKa = 4.35SWTYY419 pKa = 8.82SEE421 pKa = 5.73KK422 pKa = 10.09MFFYY426 pKa = 10.67GSEE429 pKa = 4.35TPVDD433 pKa = 4.11LMPTSGGAIYY443 pKa = 10.38HH444 pKa = 6.14GFFTGASDD452 pKa = 3.84DD453 pKa = 3.67VDD455 pKa = 4.19YY456 pKa = 11.19GWEE459 pKa = 3.96EE460 pKa = 3.9FFGDD464 pKa = 5.03FSLAVNWEE472 pKa = 3.95TGKK475 pKa = 10.41VVGYY479 pKa = 10.17FIDD482 pKa = 4.25DD483 pKa = 3.41EE484 pKa = 5.07HH485 pKa = 8.35YY486 pKa = 10.5GRR488 pKa = 11.84PIPVYY493 pKa = 9.71LTGEE497 pKa = 4.08VDD499 pKa = 3.83GSSLSSVSIFGLDD512 pKa = 3.21YY513 pKa = 11.29DD514 pKa = 5.02DD515 pKa = 5.39YY516 pKa = 11.87GDD518 pKa = 4.11GALYY522 pKa = 10.11DD523 pKa = 3.46AWITTGSGTFAQFYY537 pKa = 7.12GTYY540 pKa = 8.79YY541 pKa = 10.37QGLGLVAEE549 pKa = 4.85GQSGSLYY556 pKa = 10.66YY557 pKa = 10.77DD558 pKa = 4.2DD559 pKa = 5.85AQEE562 pKa = 4.15DD563 pKa = 4.08WSVVGGGFRR572 pKa = 11.84DD573 pKa = 3.7PVFTLPEE580 pKa = 3.86QAASSTWRR588 pKa = 11.84GFVTAMEE595 pKa = 4.45INLVEE600 pKa = 4.4EE601 pKa = 4.86PGTPPEE607 pKa = 4.3LFRR610 pKa = 11.84STSAEE615 pKa = 3.98YY616 pKa = 10.74FEE618 pKa = 5.33LQIDD622 pKa = 4.1RR623 pKa = 11.84QAGMFSGQIIEE634 pKa = 4.76AEE636 pKa = 4.36SATTTGLSIGGLEE649 pKa = 3.94VAGFYY654 pKa = 11.08VSDD657 pKa = 3.6NFLAGEE663 pKa = 4.56LSSASDD669 pKa = 3.45SDD671 pKa = 3.79LQGALGGSGEE681 pKa = 4.35YY682 pKa = 10.63YY683 pKa = 10.4FYY685 pKa = 11.22DD686 pKa = 3.97FEE688 pKa = 6.94DD689 pKa = 3.84EE690 pKa = 4.42ADD692 pKa = 3.64YY693 pKa = 10.7FQIGEE698 pKa = 4.25LAEE701 pKa = 4.26HH702 pKa = 6.19VSWGYY707 pKa = 9.68WGAAFTGQDD716 pKa = 3.46EE717 pKa = 4.55MPYY720 pKa = 9.93LISPAGSFWVAGEE733 pKa = 4.11LTPADD738 pKa = 4.09TVLGMIDD745 pKa = 3.73AEE747 pKa = 4.48VTGTYY752 pKa = 10.16EE753 pKa = 4.2GGAEE757 pKa = 4.38GILIFEE763 pKa = 4.48GTMADD768 pKa = 4.41LYY770 pKa = 11.35NGQTTLNINFGDD782 pKa = 3.78ANINGSITFSEE793 pKa = 4.38IAEE796 pKa = 4.13LTIDD800 pKa = 4.15GSSSLGPDD808 pKa = 3.56GFAADD813 pKa = 3.68VTSVEE818 pKa = 4.33FFGGEE823 pKa = 3.74PTLASGGISGAFFGPNAEE841 pKa = 4.13SVGGNFHH848 pKa = 7.25AEE850 pKa = 3.64GGGAQFLGIFGGDD863 pKa = 2.89RR864 pKa = 3.28
MM1 pKa = 7.18SQRR4 pKa = 11.84PLLQPVVFLAALVCLLAVTFWSAPPATAAEE34 pKa = 4.32EE35 pKa = 4.36VATVVALRR43 pKa = 11.84GTAVAVDD50 pKa = 3.19AAGAEE55 pKa = 3.82RR56 pKa = 11.84ALAIKK61 pKa = 10.48APIHH65 pKa = 5.65RR66 pKa = 11.84QDD68 pKa = 3.83TIRR71 pKa = 11.84TGPRR75 pKa = 11.84GRR77 pKa = 11.84LQLMFTDD84 pKa = 3.63NTIISLGVNAEE95 pKa = 4.16LEE97 pKa = 4.02IAEE100 pKa = 4.4YY101 pKa = 10.56QWDD104 pKa = 3.86QQEE107 pKa = 4.82GEE109 pKa = 4.22LNTRR113 pKa = 11.84VNQGAFRR120 pKa = 11.84VMGGSVARR128 pKa = 11.84TSPEE132 pKa = 3.64KK133 pKa = 10.28FTTDD137 pKa = 2.92TPAATIGIRR146 pKa = 11.84GSMYY150 pKa = 10.44AGRR153 pKa = 11.84VSDD156 pKa = 3.68GALAVVFQGGTGIIVSNPAGTVEE179 pKa = 3.81ITTPGMGTRR188 pKa = 11.84VASRR192 pKa = 11.84TAAPTPPTVFTQEE205 pKa = 4.06DD206 pKa = 3.9LGALEE211 pKa = 5.0DD212 pKa = 4.06EE213 pKa = 4.84EE214 pKa = 4.74VDD216 pKa = 3.51EE217 pKa = 4.6EE218 pKa = 4.2AAEE221 pKa = 4.18EE222 pKa = 4.15EE223 pKa = 4.56TVEE226 pKa = 4.7DD227 pKa = 3.8EE228 pKa = 4.73TPAEE232 pKa = 4.18EE233 pKa = 4.16PAEE236 pKa = 4.24EE237 pKa = 4.19EE238 pKa = 4.34WEE240 pKa = 4.13DD241 pKa = 3.53AALDD245 pKa = 4.03EE246 pKa = 5.0PEE248 pKa = 4.09EE249 pKa = 4.11AAAPVAAAPAPITDD263 pKa = 3.8TAVDD267 pKa = 3.64QATEE271 pKa = 4.24TTTDD275 pKa = 3.23AAQDD279 pKa = 3.25EE280 pKa = 4.78WEE282 pKa = 4.28QTVVEE287 pKa = 4.81ADD289 pKa = 4.16PEE291 pKa = 4.31PTDD294 pKa = 4.65PDD296 pKa = 3.72PTDD299 pKa = 4.37PEE301 pKa = 5.56PIDD304 pKa = 4.6PEE306 pKa = 4.05PTDD309 pKa = 3.66PSEE312 pKa = 3.88AVYY315 pKa = 10.33SGRR318 pKa = 11.84GIEE321 pKa = 4.44IIYY324 pKa = 10.58SSMQEE329 pKa = 4.01EE330 pKa = 4.81VAVEE334 pKa = 3.93HH335 pKa = 6.82LVLTDD340 pKa = 3.82KK341 pKa = 10.9PSEE344 pKa = 4.02LTEE347 pKa = 4.14IEE349 pKa = 4.45AGLLEE354 pKa = 4.39YY355 pKa = 10.97NEE357 pKa = 4.32QSDD360 pKa = 4.17FSDD363 pKa = 3.92GEE365 pKa = 4.34YY366 pKa = 10.53SAVWSYY372 pKa = 11.82TDD374 pKa = 3.27ATQVEE379 pKa = 4.87EE380 pKa = 4.48WEE382 pKa = 4.51EE383 pKa = 4.27GDD385 pKa = 5.54DD386 pKa = 5.09FDD388 pKa = 5.32FQFTFVMVDD397 pKa = 3.21DD398 pKa = 5.35RR399 pKa = 11.84GEE401 pKa = 3.81FWLYY405 pKa = 8.31EE406 pKa = 3.92QWSDD410 pKa = 4.08TEE412 pKa = 4.37TDD414 pKa = 3.56EE415 pKa = 4.35SWTYY419 pKa = 8.82SEE421 pKa = 5.73KK422 pKa = 10.09MFFYY426 pKa = 10.67GSEE429 pKa = 4.35TPVDD433 pKa = 4.11LMPTSGGAIYY443 pKa = 10.38HH444 pKa = 6.14GFFTGASDD452 pKa = 3.84DD453 pKa = 3.67VDD455 pKa = 4.19YY456 pKa = 11.19GWEE459 pKa = 3.96EE460 pKa = 3.9FFGDD464 pKa = 5.03FSLAVNWEE472 pKa = 3.95TGKK475 pKa = 10.41VVGYY479 pKa = 10.17FIDD482 pKa = 4.25DD483 pKa = 3.41EE484 pKa = 5.07HH485 pKa = 8.35YY486 pKa = 10.5GRR488 pKa = 11.84PIPVYY493 pKa = 9.71LTGEE497 pKa = 4.08VDD499 pKa = 3.83GSSLSSVSIFGLDD512 pKa = 3.21YY513 pKa = 11.29DD514 pKa = 5.02DD515 pKa = 5.39YY516 pKa = 11.87GDD518 pKa = 4.11GALYY522 pKa = 10.11DD523 pKa = 3.46AWITTGSGTFAQFYY537 pKa = 7.12GTYY540 pKa = 8.79YY541 pKa = 10.37QGLGLVAEE549 pKa = 4.85GQSGSLYY556 pKa = 10.66YY557 pKa = 10.77DD558 pKa = 4.2DD559 pKa = 5.85AQEE562 pKa = 4.15DD563 pKa = 4.08WSVVGGGFRR572 pKa = 11.84DD573 pKa = 3.7PVFTLPEE580 pKa = 3.86QAASSTWRR588 pKa = 11.84GFVTAMEE595 pKa = 4.45INLVEE600 pKa = 4.4EE601 pKa = 4.86PGTPPEE607 pKa = 4.3LFRR610 pKa = 11.84STSAEE615 pKa = 3.98YY616 pKa = 10.74FEE618 pKa = 5.33LQIDD622 pKa = 4.1RR623 pKa = 11.84QAGMFSGQIIEE634 pKa = 4.76AEE636 pKa = 4.36SATTTGLSIGGLEE649 pKa = 3.94VAGFYY654 pKa = 11.08VSDD657 pKa = 3.6NFLAGEE663 pKa = 4.56LSSASDD669 pKa = 3.45SDD671 pKa = 3.79LQGALGGSGEE681 pKa = 4.35YY682 pKa = 10.63YY683 pKa = 10.4FYY685 pKa = 11.22DD686 pKa = 3.97FEE688 pKa = 6.94DD689 pKa = 3.84EE690 pKa = 4.42ADD692 pKa = 3.64YY693 pKa = 10.7FQIGEE698 pKa = 4.25LAEE701 pKa = 4.26HH702 pKa = 6.19VSWGYY707 pKa = 9.68WGAAFTGQDD716 pKa = 3.46EE717 pKa = 4.55MPYY720 pKa = 9.93LISPAGSFWVAGEE733 pKa = 4.11LTPADD738 pKa = 4.09TVLGMIDD745 pKa = 3.73AEE747 pKa = 4.48VTGTYY752 pKa = 10.16EE753 pKa = 4.2GGAEE757 pKa = 4.38GILIFEE763 pKa = 4.48GTMADD768 pKa = 4.41LYY770 pKa = 11.35NGQTTLNINFGDD782 pKa = 3.78ANINGSITFSEE793 pKa = 4.38IAEE796 pKa = 4.13LTIDD800 pKa = 4.15GSSSLGPDD808 pKa = 3.56GFAADD813 pKa = 3.68VTSVEE818 pKa = 4.33FFGGEE823 pKa = 3.74PTLASGGISGAFFGPNAEE841 pKa = 4.13SVGGNFHH848 pKa = 7.25AEE850 pKa = 3.64GGGAQFLGIFGGDD863 pKa = 2.89RR864 pKa = 3.28
Molecular weight: 92.64 kDa
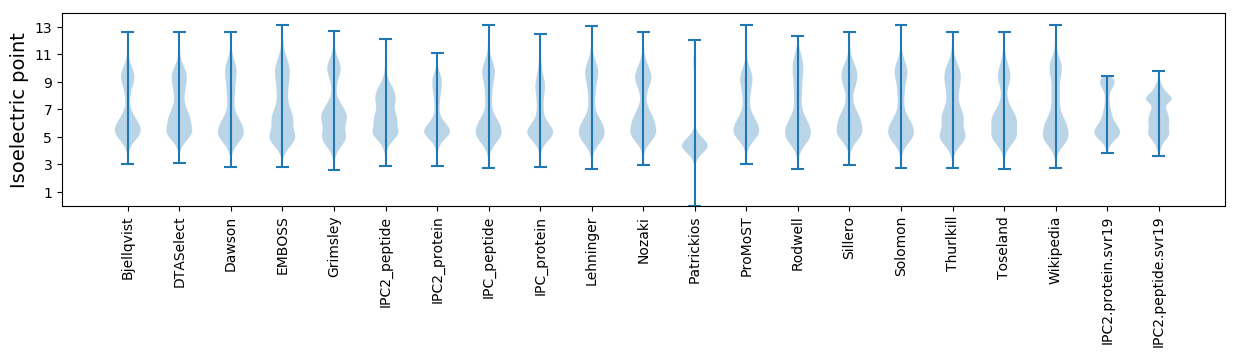
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D6Z0V9|D6Z0V9_DESAT Putative membrane protein insertion efficiency factor OS=Desulfurivibrio alkaliphilus (strain DSM 19089 / UNIQEM U267 / AHT2) OX=589865 GN=DaAHT2_2555 PE=3 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.22QPSNTKK12 pKa = 8.42RR13 pKa = 11.84HH14 pKa = 5.05RR15 pKa = 11.84THH17 pKa = 6.71GFRR20 pKa = 11.84VRR22 pKa = 11.84MSTKK26 pKa = 10.09NGRR29 pKa = 11.84AVINRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.58GRR40 pKa = 11.84KK41 pKa = 8.66RR42 pKa = 11.84LAVV45 pKa = 3.41
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.22QPSNTKK12 pKa = 8.42RR13 pKa = 11.84HH14 pKa = 5.05RR15 pKa = 11.84THH17 pKa = 6.71GFRR20 pKa = 11.84VRR22 pKa = 11.84MSTKK26 pKa = 10.09NGRR29 pKa = 11.84AVINRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.58GRR40 pKa = 11.84KK41 pKa = 8.66RR42 pKa = 11.84LAVV45 pKa = 3.41
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
903674 |
30 |
2439 |
348.2 |
38.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.575 ± 0.06 | 1.186 ± 0.025 |
5.138 ± 0.033 | 6.861 ± 0.046 |
3.674 ± 0.022 | 7.965 ± 0.043 |
2.269 ± 0.026 | 5.096 ± 0.035 |
3.684 ± 0.044 | 11.524 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.271 ± 0.019 | 2.951 ± 0.033 |
5.239 ± 0.038 | 4.172 ± 0.032 |
7.255 ± 0.045 | 4.895 ± 0.032 |
4.696 ± 0.03 | 6.791 ± 0.04 |
1.142 ± 0.017 | 2.614 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
