
Streptococcus henryi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
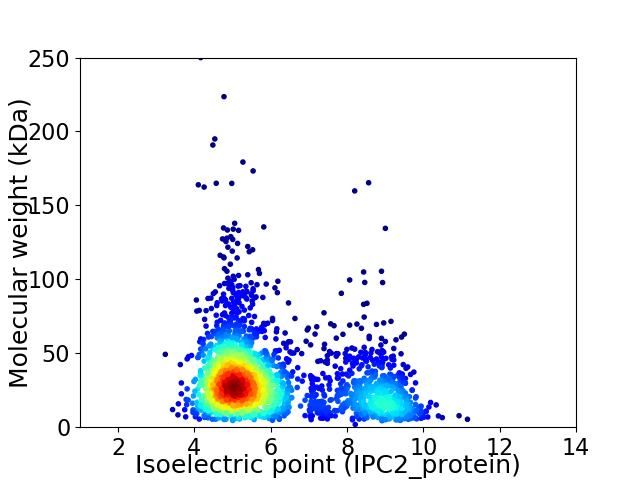
Virtual 2D-PAGE plot for 2315 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G6BDD2|A0A1G6BDD2_9STRE GyrI-like domain-containing protein OS=Streptococcus henryi OX=439219 GN=SAMN02910293_00929 PE=4 SV=1
MM1 pKa = 7.42IKK3 pKa = 10.17VKK5 pKa = 10.45KK6 pKa = 10.59VLVLLFSVFLLASCSSNKK24 pKa = 9.86SSEE27 pKa = 3.91NAIDD31 pKa = 3.49EE32 pKa = 4.48GQIEE36 pKa = 4.85LIKK39 pKa = 10.66DD40 pKa = 3.45VQASVSEE47 pKa = 4.01NFYY50 pKa = 10.41YY51 pKa = 10.37QQLSDD56 pKa = 3.92VEE58 pKa = 4.32KK59 pKa = 10.73EE60 pKa = 3.7NYY62 pKa = 9.63ARR64 pKa = 11.84LRR66 pKa = 11.84LAFNDD71 pKa = 4.13FSDD74 pKa = 3.73EE75 pKa = 4.18VTLKK79 pKa = 10.81EE80 pKa = 4.46VDD82 pKa = 3.98DD83 pKa = 4.51EE84 pKa = 4.58SLAKK88 pKa = 10.35ISTAFTNDD96 pKa = 3.14NPDD99 pKa = 3.39VFWFNDD105 pKa = 3.14YY106 pKa = 10.2TYY108 pKa = 9.72QTDD111 pKa = 4.21GNDD114 pKa = 3.38QLILEE119 pKa = 5.0IEE121 pKa = 4.33TPDD124 pKa = 3.67DD125 pKa = 3.55VEE127 pKa = 5.02EE128 pKa = 4.49IYY130 pKa = 10.9QLIEE134 pKa = 4.05VMSEE138 pKa = 3.81QILAEE143 pKa = 4.26MPTDD147 pKa = 3.37SDD149 pKa = 4.01YY150 pKa = 11.94SRR152 pKa = 11.84VKK154 pKa = 10.5YY155 pKa = 10.35LYY157 pKa = 10.3EE158 pKa = 4.31FIINYY163 pKa = 8.7TDD165 pKa = 3.65YY166 pKa = 10.12NTLALTDD173 pKa = 4.42DD174 pKa = 4.14SEE176 pKa = 4.53LDD178 pKa = 3.08KK179 pKa = 11.65GQDD182 pKa = 3.05IRR184 pKa = 11.84SVFIDD189 pKa = 3.46QLSVCSGYY197 pKa = 11.21SKK199 pKa = 10.95AFQYY203 pKa = 10.54LANKK207 pKa = 10.0AGIPTTYY214 pKa = 9.67IVGFARR220 pKa = 11.84SDD222 pKa = 3.27KK223 pKa = 10.44TEE225 pKa = 3.97EE226 pKa = 4.27SEE228 pKa = 6.38LPHH231 pKa = 6.49AWNTVIINGQSYY243 pKa = 10.57NVDD246 pKa = 3.85TTWGDD251 pKa = 3.72PIFEE255 pKa = 4.7GEE257 pKa = 4.1MSEE260 pKa = 5.74DD261 pKa = 3.28MSDD264 pKa = 3.58TINYY268 pKa = 8.8HH269 pKa = 5.76FLCVPDD275 pKa = 3.77YY276 pKa = 11.27LFNNSHH282 pKa = 5.95TASDD286 pKa = 5.13DD287 pKa = 3.33ILFQDD292 pKa = 4.78GSSIDD297 pKa = 4.47DD298 pKa = 3.45VWTLPTSTDD307 pKa = 2.83NSLNYY312 pKa = 9.67YY313 pKa = 8.62VQAGCFFEE321 pKa = 5.05SYY323 pKa = 9.83QRR325 pKa = 11.84GEE327 pKa = 3.9MEE329 pKa = 4.59SYY331 pKa = 10.78VEE333 pKa = 3.94QLSHH337 pKa = 7.18SEE339 pKa = 3.79NLKK342 pKa = 10.17QIEE345 pKa = 4.32LVFEE349 pKa = 4.21NQEE352 pKa = 4.21SYY354 pKa = 11.66DD355 pKa = 3.71LALNDD360 pKa = 4.09LLEE363 pKa = 4.43EE364 pKa = 4.56GFFGEE369 pKa = 5.72LIQKK373 pKa = 9.86YY374 pKa = 9.83ILGDD378 pKa = 2.96IDD380 pKa = 3.9FEE382 pKa = 5.43YY383 pKa = 11.31NFIYY387 pKa = 10.69DD388 pKa = 3.55DD389 pKa = 3.73VNYY392 pKa = 9.2YY393 pKa = 9.98ICINIPGRR401 pKa = 3.61
MM1 pKa = 7.42IKK3 pKa = 10.17VKK5 pKa = 10.45KK6 pKa = 10.59VLVLLFSVFLLASCSSNKK24 pKa = 9.86SSEE27 pKa = 3.91NAIDD31 pKa = 3.49EE32 pKa = 4.48GQIEE36 pKa = 4.85LIKK39 pKa = 10.66DD40 pKa = 3.45VQASVSEE47 pKa = 4.01NFYY50 pKa = 10.41YY51 pKa = 10.37QQLSDD56 pKa = 3.92VEE58 pKa = 4.32KK59 pKa = 10.73EE60 pKa = 3.7NYY62 pKa = 9.63ARR64 pKa = 11.84LRR66 pKa = 11.84LAFNDD71 pKa = 4.13FSDD74 pKa = 3.73EE75 pKa = 4.18VTLKK79 pKa = 10.81EE80 pKa = 4.46VDD82 pKa = 3.98DD83 pKa = 4.51EE84 pKa = 4.58SLAKK88 pKa = 10.35ISTAFTNDD96 pKa = 3.14NPDD99 pKa = 3.39VFWFNDD105 pKa = 3.14YY106 pKa = 10.2TYY108 pKa = 9.72QTDD111 pKa = 4.21GNDD114 pKa = 3.38QLILEE119 pKa = 5.0IEE121 pKa = 4.33TPDD124 pKa = 3.67DD125 pKa = 3.55VEE127 pKa = 5.02EE128 pKa = 4.49IYY130 pKa = 10.9QLIEE134 pKa = 4.05VMSEE138 pKa = 3.81QILAEE143 pKa = 4.26MPTDD147 pKa = 3.37SDD149 pKa = 4.01YY150 pKa = 11.94SRR152 pKa = 11.84VKK154 pKa = 10.5YY155 pKa = 10.35LYY157 pKa = 10.3EE158 pKa = 4.31FIINYY163 pKa = 8.7TDD165 pKa = 3.65YY166 pKa = 10.12NTLALTDD173 pKa = 4.42DD174 pKa = 4.14SEE176 pKa = 4.53LDD178 pKa = 3.08KK179 pKa = 11.65GQDD182 pKa = 3.05IRR184 pKa = 11.84SVFIDD189 pKa = 3.46QLSVCSGYY197 pKa = 11.21SKK199 pKa = 10.95AFQYY203 pKa = 10.54LANKK207 pKa = 10.0AGIPTTYY214 pKa = 9.67IVGFARR220 pKa = 11.84SDD222 pKa = 3.27KK223 pKa = 10.44TEE225 pKa = 3.97EE226 pKa = 4.27SEE228 pKa = 6.38LPHH231 pKa = 6.49AWNTVIINGQSYY243 pKa = 10.57NVDD246 pKa = 3.85TTWGDD251 pKa = 3.72PIFEE255 pKa = 4.7GEE257 pKa = 4.1MSEE260 pKa = 5.74DD261 pKa = 3.28MSDD264 pKa = 3.58TINYY268 pKa = 8.8HH269 pKa = 5.76FLCVPDD275 pKa = 3.77YY276 pKa = 11.27LFNNSHH282 pKa = 5.95TASDD286 pKa = 5.13DD287 pKa = 3.33ILFQDD292 pKa = 4.78GSSIDD297 pKa = 4.47DD298 pKa = 3.45VWTLPTSTDD307 pKa = 2.83NSLNYY312 pKa = 9.67YY313 pKa = 8.62VQAGCFFEE321 pKa = 5.05SYY323 pKa = 9.83QRR325 pKa = 11.84GEE327 pKa = 3.9MEE329 pKa = 4.59SYY331 pKa = 10.78VEE333 pKa = 3.94QLSHH337 pKa = 7.18SEE339 pKa = 3.79NLKK342 pKa = 10.17QIEE345 pKa = 4.32LVFEE349 pKa = 4.21NQEE352 pKa = 4.21SYY354 pKa = 11.66DD355 pKa = 3.71LALNDD360 pKa = 4.09LLEE363 pKa = 4.43EE364 pKa = 4.56GFFGEE369 pKa = 5.72LIQKK373 pKa = 9.86YY374 pKa = 9.83ILGDD378 pKa = 2.96IDD380 pKa = 3.9FEE382 pKa = 5.43YY383 pKa = 11.31NFIYY387 pKa = 10.69DD388 pKa = 3.55DD389 pKa = 3.73VNYY392 pKa = 9.2YY393 pKa = 9.98ICINIPGRR401 pKa = 3.61
Molecular weight: 46.17 kDa
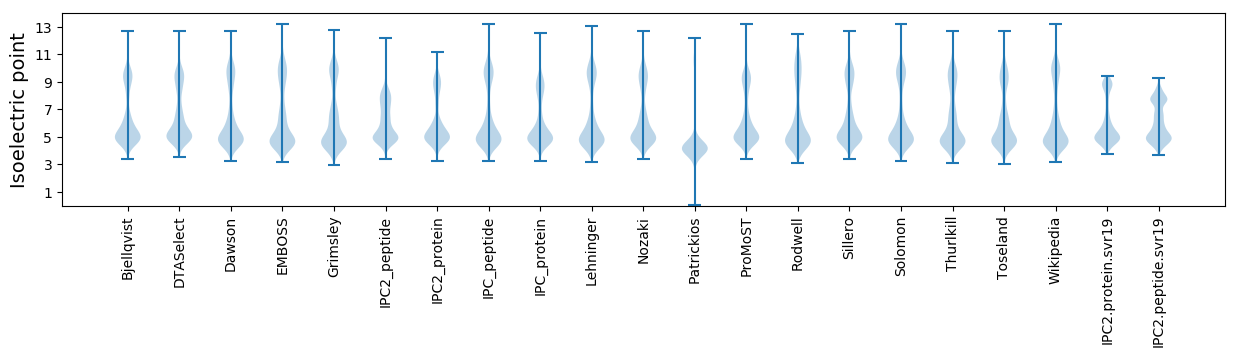
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G6D2V2|A0A1G6D2V2_9STRE Transposase IS200 like (Fragment) OS=Streptococcus henryi OX=439219 GN=SAMN02910293_01943 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.67IRR11 pKa = 11.84RR12 pKa = 11.84QRR14 pKa = 11.84KK15 pKa = 7.52HH16 pKa = 5.2GFRR19 pKa = 11.84HH20 pKa = 6.39RR21 pKa = 11.84MSTKK25 pKa = 9.02NGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.67IRR11 pKa = 11.84RR12 pKa = 11.84QRR14 pKa = 11.84KK15 pKa = 7.52HH16 pKa = 5.2GFRR19 pKa = 11.84HH20 pKa = 6.39RR21 pKa = 11.84MSTKK25 pKa = 9.02NGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
692190 |
15 |
2261 |
299.0 |
33.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.503 ± 0.062 | 0.531 ± 0.012 |
6.059 ± 0.048 | 6.989 ± 0.055 |
4.615 ± 0.045 | 6.483 ± 0.046 |
1.77 ± 0.023 | 7.299 ± 0.053 |
6.789 ± 0.046 | 9.848 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.481 ± 0.028 | 4.745 ± 0.04 |
3.155 ± 0.028 | 3.917 ± 0.033 |
3.853 ± 0.037 | 6.486 ± 0.066 |
5.79 ± 0.041 | 6.877 ± 0.043 |
0.907 ± 0.017 | 3.904 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
