
Paenibacillus sp. 597
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Paenibacillus; unclassified Paenibacillus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
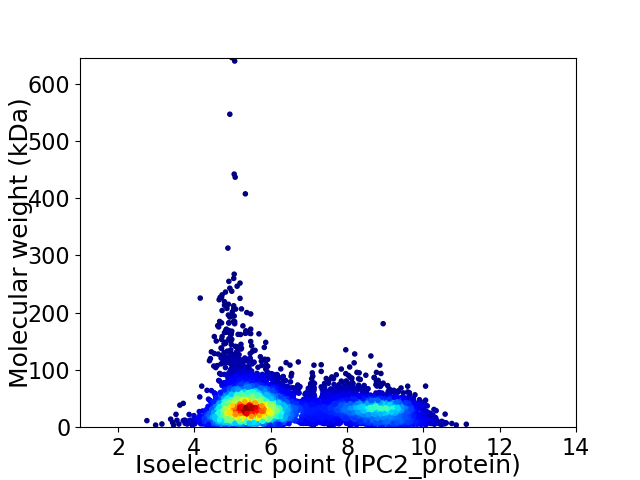
Virtual 2D-PAGE plot for 7604 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A561Q655|A0A561Q655_9BACL 2-dehydro-3-deoxygluconokinase OS=Paenibacillus sp. 597 OX=2587047 GN=FHW43_104103 PE=4 SV=1
MM1 pKa = 7.93ADD3 pKa = 2.88MMLRR7 pKa = 11.84PDD9 pKa = 4.15LKK11 pKa = 9.01TAGGEE16 pKa = 4.3VCDD19 pKa = 3.76IMYY22 pKa = 10.58NGEE25 pKa = 4.21FVGTLTLVYY34 pKa = 10.26RR35 pKa = 11.84EE36 pKa = 3.96SDD38 pKa = 3.24RR39 pKa = 11.84LKK41 pKa = 11.19GSIHH45 pKa = 7.29LEE47 pKa = 3.93RR48 pKa = 11.84QSIAQEE54 pKa = 4.22DD55 pKa = 4.34KK56 pKa = 8.73EE57 pKa = 4.48SVYY60 pKa = 11.32AFVQSYY66 pKa = 8.29IQSVIDD72 pKa = 3.51ALAIEE77 pKa = 4.48EE78 pKa = 4.69CEE80 pKa = 4.36VVANYY85 pKa = 10.66SDD87 pKa = 4.01YY88 pKa = 11.7DD89 pKa = 3.57FVIATDD95 pKa = 3.73HH96 pKa = 7.17AIGPVLGEE104 pKa = 4.03EE105 pKa = 4.99DD106 pKa = 4.56DD107 pKa = 5.42RR108 pKa = 11.84SDD110 pKa = 4.14PYY112 pKa = 11.78YY113 pKa = 11.04LDD115 pKa = 6.01DD116 pKa = 5.04EE117 pKa = 4.82DD118 pKa = 6.38ADD120 pKa = 4.27YY121 pKa = 11.39EE122 pKa = 4.46WVNDD126 pKa = 3.74DD127 pKa = 4.95EE128 pKa = 5.84IRR130 pKa = 11.84FDD132 pKa = 6.6DD133 pKa = 3.9IDD135 pKa = 3.75GTNGPEE141 pKa = 3.85SLYY144 pKa = 10.32MIEE147 pKa = 5.07QRR149 pKa = 11.84RR150 pKa = 11.84QQLGYY155 pKa = 9.09EE156 pKa = 3.96LVIVGEE162 pKa = 3.99YY163 pKa = 10.26RR164 pKa = 11.84NLVEE168 pKa = 3.58YY169 pKa = 10.23HH170 pKa = 6.69VYY172 pKa = 10.54DD173 pKa = 4.02KK174 pKa = 11.14EE175 pKa = 4.55SEE177 pKa = 4.31LVAEE181 pKa = 5.21AVTHH185 pKa = 5.85MYY187 pKa = 10.47GSEE190 pKa = 4.21VNGEE194 pKa = 4.23VTWRR198 pKa = 11.84FDD200 pKa = 3.63PFDD203 pKa = 3.96EE204 pKa = 4.68EE205 pKa = 4.06MDD207 pKa = 3.9LVTEE211 pKa = 5.25LIVADD216 pKa = 4.0YY217 pKa = 11.45DD218 pKa = 3.79EE219 pKa = 6.38DD220 pKa = 3.82EE221 pKa = 4.33TDD223 pKa = 3.21TFIFNMIFDD232 pKa = 4.52GEE234 pKa = 4.51VIDD237 pKa = 4.33TVEE240 pKa = 4.11LTHH243 pKa = 7.34IDD245 pKa = 3.85LLEE248 pKa = 4.23LVDD251 pKa = 5.64DD252 pKa = 4.05IRR254 pKa = 11.84EE255 pKa = 4.01EE256 pKa = 4.35SLMPPDD262 pKa = 4.6PEE264 pKa = 6.62DD265 pKa = 3.45YY266 pKa = 10.55TITLARR272 pKa = 11.84DD273 pKa = 3.6DD274 pKa = 5.11GDD276 pKa = 3.53TLTYY280 pKa = 10.34EE281 pKa = 5.21IYY283 pKa = 8.77QQSYY287 pKa = 8.98GGLPIGTATVDD298 pKa = 2.86ISSRR302 pKa = 11.84QLTGFIDD309 pKa = 3.9FRR311 pKa = 11.84EE312 pKa = 4.54PGDD315 pKa = 3.53SDD317 pKa = 4.02DD318 pKa = 5.17RR319 pKa = 11.84EE320 pKa = 4.47LIAALLLDD328 pKa = 4.65EE329 pKa = 5.37LDD331 pKa = 4.27KK332 pKa = 11.75EE333 pKa = 4.21NDD335 pKa = 3.45YY336 pKa = 10.02EE337 pKa = 4.37TFNVSMLYY345 pKa = 10.0RR346 pKa = 11.84NKK348 pKa = 10.64LIDD351 pKa = 4.89EE352 pKa = 4.25IWFEE356 pKa = 4.17TDD358 pKa = 2.74QLHH361 pKa = 6.37
MM1 pKa = 7.93ADD3 pKa = 2.88MMLRR7 pKa = 11.84PDD9 pKa = 4.15LKK11 pKa = 9.01TAGGEE16 pKa = 4.3VCDD19 pKa = 3.76IMYY22 pKa = 10.58NGEE25 pKa = 4.21FVGTLTLVYY34 pKa = 10.26RR35 pKa = 11.84EE36 pKa = 3.96SDD38 pKa = 3.24RR39 pKa = 11.84LKK41 pKa = 11.19GSIHH45 pKa = 7.29LEE47 pKa = 3.93RR48 pKa = 11.84QSIAQEE54 pKa = 4.22DD55 pKa = 4.34KK56 pKa = 8.73EE57 pKa = 4.48SVYY60 pKa = 11.32AFVQSYY66 pKa = 8.29IQSVIDD72 pKa = 3.51ALAIEE77 pKa = 4.48EE78 pKa = 4.69CEE80 pKa = 4.36VVANYY85 pKa = 10.66SDD87 pKa = 4.01YY88 pKa = 11.7DD89 pKa = 3.57FVIATDD95 pKa = 3.73HH96 pKa = 7.17AIGPVLGEE104 pKa = 4.03EE105 pKa = 4.99DD106 pKa = 4.56DD107 pKa = 5.42RR108 pKa = 11.84SDD110 pKa = 4.14PYY112 pKa = 11.78YY113 pKa = 11.04LDD115 pKa = 6.01DD116 pKa = 5.04EE117 pKa = 4.82DD118 pKa = 6.38ADD120 pKa = 4.27YY121 pKa = 11.39EE122 pKa = 4.46WVNDD126 pKa = 3.74DD127 pKa = 4.95EE128 pKa = 5.84IRR130 pKa = 11.84FDD132 pKa = 6.6DD133 pKa = 3.9IDD135 pKa = 3.75GTNGPEE141 pKa = 3.85SLYY144 pKa = 10.32MIEE147 pKa = 5.07QRR149 pKa = 11.84RR150 pKa = 11.84QQLGYY155 pKa = 9.09EE156 pKa = 3.96LVIVGEE162 pKa = 3.99YY163 pKa = 10.26RR164 pKa = 11.84NLVEE168 pKa = 3.58YY169 pKa = 10.23HH170 pKa = 6.69VYY172 pKa = 10.54DD173 pKa = 4.02KK174 pKa = 11.14EE175 pKa = 4.55SEE177 pKa = 4.31LVAEE181 pKa = 5.21AVTHH185 pKa = 5.85MYY187 pKa = 10.47GSEE190 pKa = 4.21VNGEE194 pKa = 4.23VTWRR198 pKa = 11.84FDD200 pKa = 3.63PFDD203 pKa = 3.96EE204 pKa = 4.68EE205 pKa = 4.06MDD207 pKa = 3.9LVTEE211 pKa = 5.25LIVADD216 pKa = 4.0YY217 pKa = 11.45DD218 pKa = 3.79EE219 pKa = 6.38DD220 pKa = 3.82EE221 pKa = 4.33TDD223 pKa = 3.21TFIFNMIFDD232 pKa = 4.52GEE234 pKa = 4.51VIDD237 pKa = 4.33TVEE240 pKa = 4.11LTHH243 pKa = 7.34IDD245 pKa = 3.85LLEE248 pKa = 4.23LVDD251 pKa = 5.64DD252 pKa = 4.05IRR254 pKa = 11.84EE255 pKa = 4.01EE256 pKa = 4.35SLMPPDD262 pKa = 4.6PEE264 pKa = 6.62DD265 pKa = 3.45YY266 pKa = 10.55TITLARR272 pKa = 11.84DD273 pKa = 3.6DD274 pKa = 5.11GDD276 pKa = 3.53TLTYY280 pKa = 10.34EE281 pKa = 5.21IYY283 pKa = 8.77QQSYY287 pKa = 8.98GGLPIGTATVDD298 pKa = 2.86ISSRR302 pKa = 11.84QLTGFIDD309 pKa = 3.9FRR311 pKa = 11.84EE312 pKa = 4.54PGDD315 pKa = 3.53SDD317 pKa = 4.02DD318 pKa = 5.17RR319 pKa = 11.84EE320 pKa = 4.47LIAALLLDD328 pKa = 4.65EE329 pKa = 5.37LDD331 pKa = 4.27KK332 pKa = 11.75EE333 pKa = 4.21NDD335 pKa = 3.45YY336 pKa = 10.02EE337 pKa = 4.37TFNVSMLYY345 pKa = 10.0RR346 pKa = 11.84NKK348 pKa = 10.64LIDD351 pKa = 4.89EE352 pKa = 4.25IWFEE356 pKa = 4.17TDD358 pKa = 2.74QLHH361 pKa = 6.37
Molecular weight: 41.61 kDa
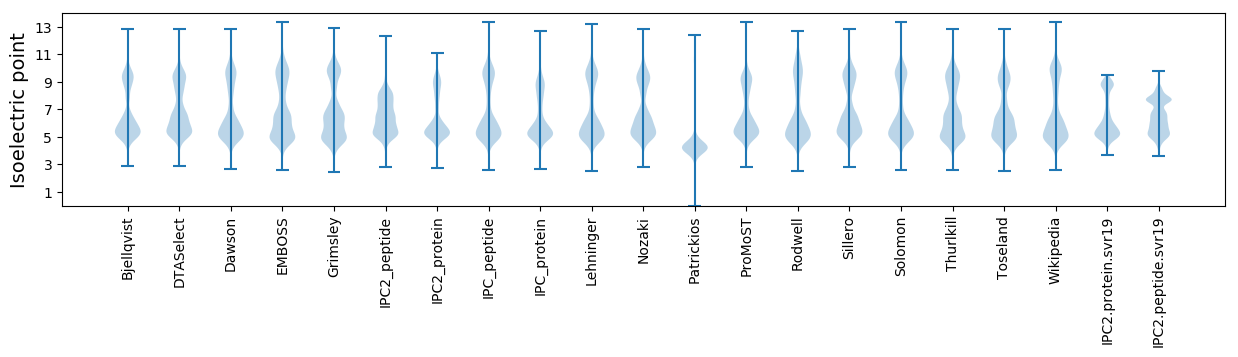
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A561PPL5|A0A561PPL5_9BACL Glycosyl transferase family 2 OS=Paenibacillus sp. 597 OX=2587047 GN=FHW43_1096 PE=4 SV=1
MM1 pKa = 7.61GPTFNPNTRR10 pKa = 11.84KK11 pKa = 9.86RR12 pKa = 11.84KK13 pKa = 8.79KK14 pKa = 9.13NHH16 pKa = 5.06GFRR19 pKa = 11.84KK20 pKa = 9.98RR21 pKa = 11.84MSTKK25 pKa = 10.0NGRR28 pKa = 11.84KK29 pKa = 8.8VLQARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.45GRR39 pKa = 11.84KK40 pKa = 8.72ILSAA44 pKa = 4.02
MM1 pKa = 7.61GPTFNPNTRR10 pKa = 11.84KK11 pKa = 9.86RR12 pKa = 11.84KK13 pKa = 8.79KK14 pKa = 9.13NHH16 pKa = 5.06GFRR19 pKa = 11.84KK20 pKa = 9.98RR21 pKa = 11.84MSTKK25 pKa = 10.0NGRR28 pKa = 11.84KK29 pKa = 8.8VLQARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.45GRR39 pKa = 11.84KK40 pKa = 8.72ILSAA44 pKa = 4.02
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2559972 |
29 |
5752 |
336.7 |
37.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.945 ± 0.031 | 0.733 ± 0.009 |
5.091 ± 0.02 | 6.282 ± 0.036 |
4.275 ± 0.02 | 7.099 ± 0.031 |
2.165 ± 0.015 | 6.959 ± 0.028 |
5.638 ± 0.026 | 10.011 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.87 ± 0.015 | 4.111 ± 0.028 |
4.006 ± 0.018 | 4.061 ± 0.021 |
4.471 ± 0.026 | 6.592 ± 0.026 |
5.74 ± 0.031 | 7.036 ± 0.026 |
1.334 ± 0.011 | 3.581 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
