
Brevibacterium celere
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Brevibacteriaceae; Brevibacterium
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
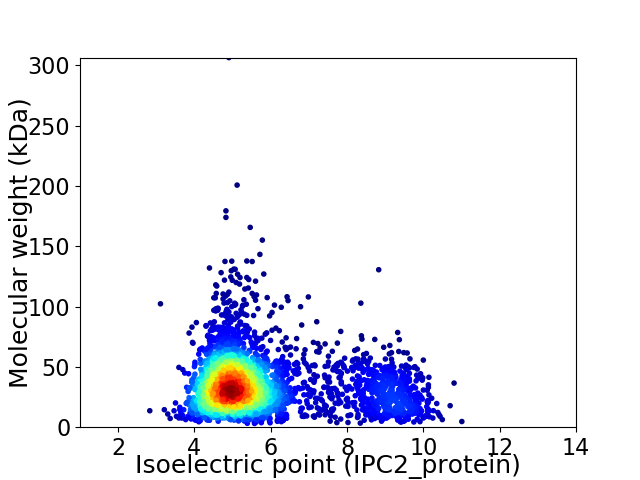
Virtual 2D-PAGE plot for 3344 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A366IR78|A0A366IR78_9MICO Mak_N_cap domain-containing protein OS=Brevibacterium celere OX=225845 GN=DFO65_101339 PE=4 SV=1
MM1 pKa = 7.47TFNPDD6 pKa = 2.96ADD8 pKa = 3.29ISGNTTSRR16 pKa = 11.84RR17 pKa = 11.84GRR19 pKa = 11.84TIGIAGGGVGVVGLLVFLVGPLLGIDD45 pKa = 3.43VTGLVGDD52 pKa = 4.49GMLEE56 pKa = 4.34GGDD59 pKa = 3.87SGQSSGEE66 pKa = 4.09SSLLEE71 pKa = 4.51CDD73 pKa = 3.17TGEE76 pKa = 4.2DD77 pKa = 3.43ANRR80 pKa = 11.84SVDD83 pKa = 3.31CRR85 pKa = 11.84MAGAQVVLDD94 pKa = 4.93DD95 pKa = 3.89YY96 pKa = 9.12WTQHH100 pKa = 4.49VEE102 pKa = 4.42GYY104 pKa = 9.85RR105 pKa = 11.84PPQMTIVDD113 pKa = 4.27GQTSTGCGTASNAVGPFYY131 pKa = 11.0CPADD135 pKa = 3.28QGVYY139 pKa = 9.79IDD141 pKa = 4.41PAFFDD146 pKa = 3.63LMRR149 pKa = 11.84EE150 pKa = 3.95RR151 pKa = 11.84FGASAGEE158 pKa = 3.92LAQLYY163 pKa = 10.02IVGHH167 pKa = 5.52EE168 pKa = 4.22WGHH171 pKa = 6.42HH172 pKa = 4.54IQNITGTMDD181 pKa = 3.07EE182 pKa = 4.94HH183 pKa = 7.0PNNGTGPDD191 pKa = 3.67SNGVRR196 pKa = 11.84MEE198 pKa = 4.16LQADD202 pKa = 4.87CYY204 pKa = 10.76AGAWLGEE211 pKa = 4.38VTTLTDD217 pKa = 4.06DD218 pKa = 3.49NGVPYY223 pKa = 10.79LEE225 pKa = 5.4APTEE229 pKa = 4.25VQLSDD234 pKa = 3.43ALNAAFVVGDD244 pKa = 4.27DD245 pKa = 4.99HH246 pKa = 7.18IQSQSGFVNPEE257 pKa = 3.94SFTHH261 pKa = 5.78GTSEE265 pKa = 4.76QRR267 pKa = 11.84QKK269 pKa = 10.69WFTQGYY275 pKa = 5.64EE276 pKa = 4.08TGLGSCDD283 pKa = 3.11SFAVRR288 pKa = 11.84GGEE291 pKa = 3.9LL292 pKa = 3.2
MM1 pKa = 7.47TFNPDD6 pKa = 2.96ADD8 pKa = 3.29ISGNTTSRR16 pKa = 11.84RR17 pKa = 11.84GRR19 pKa = 11.84TIGIAGGGVGVVGLLVFLVGPLLGIDD45 pKa = 3.43VTGLVGDD52 pKa = 4.49GMLEE56 pKa = 4.34GGDD59 pKa = 3.87SGQSSGEE66 pKa = 4.09SSLLEE71 pKa = 4.51CDD73 pKa = 3.17TGEE76 pKa = 4.2DD77 pKa = 3.43ANRR80 pKa = 11.84SVDD83 pKa = 3.31CRR85 pKa = 11.84MAGAQVVLDD94 pKa = 4.93DD95 pKa = 3.89YY96 pKa = 9.12WTQHH100 pKa = 4.49VEE102 pKa = 4.42GYY104 pKa = 9.85RR105 pKa = 11.84PPQMTIVDD113 pKa = 4.27GQTSTGCGTASNAVGPFYY131 pKa = 11.0CPADD135 pKa = 3.28QGVYY139 pKa = 9.79IDD141 pKa = 4.41PAFFDD146 pKa = 3.63LMRR149 pKa = 11.84EE150 pKa = 3.95RR151 pKa = 11.84FGASAGEE158 pKa = 3.92LAQLYY163 pKa = 10.02IVGHH167 pKa = 5.52EE168 pKa = 4.22WGHH171 pKa = 6.42HH172 pKa = 4.54IQNITGTMDD181 pKa = 3.07EE182 pKa = 4.94HH183 pKa = 7.0PNNGTGPDD191 pKa = 3.67SNGVRR196 pKa = 11.84MEE198 pKa = 4.16LQADD202 pKa = 4.87CYY204 pKa = 10.76AGAWLGEE211 pKa = 4.38VTTLTDD217 pKa = 4.06DD218 pKa = 3.49NGVPYY223 pKa = 10.79LEE225 pKa = 5.4APTEE229 pKa = 4.25VQLSDD234 pKa = 3.43ALNAAFVVGDD244 pKa = 4.27DD245 pKa = 4.99HH246 pKa = 7.18IQSQSGFVNPEE257 pKa = 3.94SFTHH261 pKa = 5.78GTSEE265 pKa = 4.76QRR267 pKa = 11.84QKK269 pKa = 10.69WFTQGYY275 pKa = 5.64EE276 pKa = 4.08TGLGSCDD283 pKa = 3.11SFAVRR288 pKa = 11.84GGEE291 pKa = 3.9LL292 pKa = 3.2
Molecular weight: 30.81 kDa
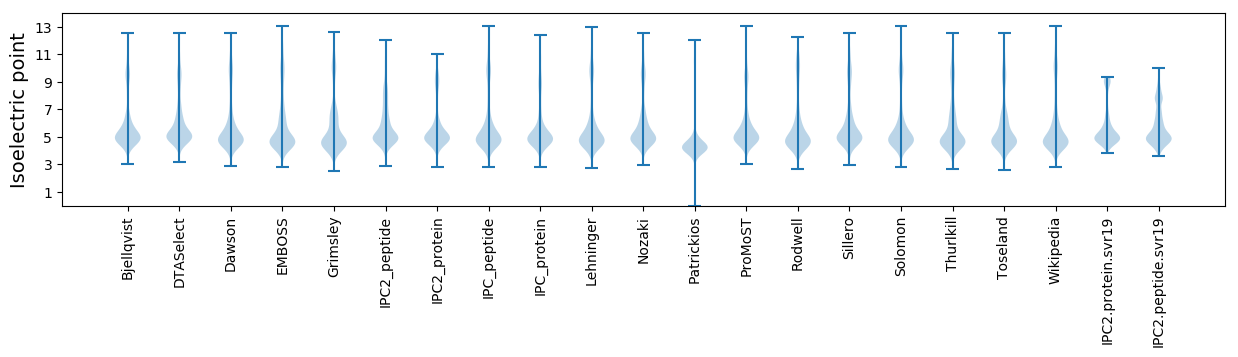
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A366ILR5|A0A366ILR5_9MICO Nitrite reductase/ring-hydroxylating ferredoxin subunit OS=Brevibacterium celere OX=225845 GN=DFO65_102195 PE=4 SV=1
MM1 pKa = 6.73TARR4 pKa = 11.84RR5 pKa = 11.84SPALPLLWAGLATIVAGAAMSLQGRR30 pKa = 11.84ANGILGPVLGHH41 pKa = 6.78AVFAALLSFATGLLLVGGALLLSARR66 pKa = 11.84SRR68 pKa = 11.84AAAVRR73 pKa = 11.84LFSLVRR79 pKa = 11.84TGALPRR85 pKa = 11.84WMLLGGCSGALVVIAQATTVPVMGIAMFTMAFVSGQVTGGLCVDD129 pKa = 4.32ATHH132 pKa = 7.2LPPGGRR138 pKa = 11.84QRR140 pKa = 11.84LTFFRR145 pKa = 11.84ILGVLVVLAALTLGAWEE162 pKa = 4.2RR163 pKa = 11.84LVLGVPLWAPLLPFASGALTALQQACNGRR192 pKa = 11.84IRR194 pKa = 11.84AATGSAVVATAVNFAVGFLLLLALTGVLLASGVRR228 pKa = 11.84WTGFPGPGQWWILLGGVLGVVFIAFTSLTVRR259 pKa = 11.84RR260 pKa = 11.84LGVLLLTLFSLFGNLAGALLLDD282 pKa = 4.62LAVPLPGSLVSTTTVLSAALVLAGIAVTLMPAAGSLRR319 pKa = 11.84GRR321 pKa = 11.84GSRR324 pKa = 11.84TRR326 pKa = 11.84GPRR329 pKa = 11.84RR330 pKa = 11.84GPSRR334 pKa = 11.84RR335 pKa = 11.84RR336 pKa = 11.84RR337 pKa = 11.84GTSRR341 pKa = 11.84GSRR344 pKa = 11.84GRR346 pKa = 11.84RR347 pKa = 11.84SAAGARR353 pKa = 11.84TRR355 pKa = 11.84SRR357 pKa = 3.25
MM1 pKa = 6.73TARR4 pKa = 11.84RR5 pKa = 11.84SPALPLLWAGLATIVAGAAMSLQGRR30 pKa = 11.84ANGILGPVLGHH41 pKa = 6.78AVFAALLSFATGLLLVGGALLLSARR66 pKa = 11.84SRR68 pKa = 11.84AAAVRR73 pKa = 11.84LFSLVRR79 pKa = 11.84TGALPRR85 pKa = 11.84WMLLGGCSGALVVIAQATTVPVMGIAMFTMAFVSGQVTGGLCVDD129 pKa = 4.32ATHH132 pKa = 7.2LPPGGRR138 pKa = 11.84QRR140 pKa = 11.84LTFFRR145 pKa = 11.84ILGVLVVLAALTLGAWEE162 pKa = 4.2RR163 pKa = 11.84LVLGVPLWAPLLPFASGALTALQQACNGRR192 pKa = 11.84IRR194 pKa = 11.84AATGSAVVATAVNFAVGFLLLLALTGVLLASGVRR228 pKa = 11.84WTGFPGPGQWWILLGGVLGVVFIAFTSLTVRR259 pKa = 11.84RR260 pKa = 11.84LGVLLLTLFSLFGNLAGALLLDD282 pKa = 4.62LAVPLPGSLVSTTTVLSAALVLAGIAVTLMPAAGSLRR319 pKa = 11.84GRR321 pKa = 11.84GSRR324 pKa = 11.84TRR326 pKa = 11.84GPRR329 pKa = 11.84RR330 pKa = 11.84GPSRR334 pKa = 11.84RR335 pKa = 11.84RR336 pKa = 11.84RR337 pKa = 11.84GTSRR341 pKa = 11.84GSRR344 pKa = 11.84GRR346 pKa = 11.84RR347 pKa = 11.84SAAGARR353 pKa = 11.84TRR355 pKa = 11.84SRR357 pKa = 3.25
Molecular weight: 36.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1135316 |
29 |
2868 |
339.5 |
36.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.534 ± 0.06 | 0.653 ± 0.01 |
6.269 ± 0.034 | 6.292 ± 0.042 |
3.229 ± 0.026 | 8.934 ± 0.04 |
2.16 ± 0.022 | 4.842 ± 0.033 |
2.186 ± 0.035 | 9.952 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.959 ± 0.015 | 1.982 ± 0.023 |
5.358 ± 0.031 | 2.603 ± 0.021 |
7.146 ± 0.042 | 6.205 ± 0.03 |
6.091 ± 0.027 | 8.383 ± 0.037 |
1.351 ± 0.014 | 1.872 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
