
Catalinimonas alkaloidigena
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Catalimonadaceae; Catalinimonas
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
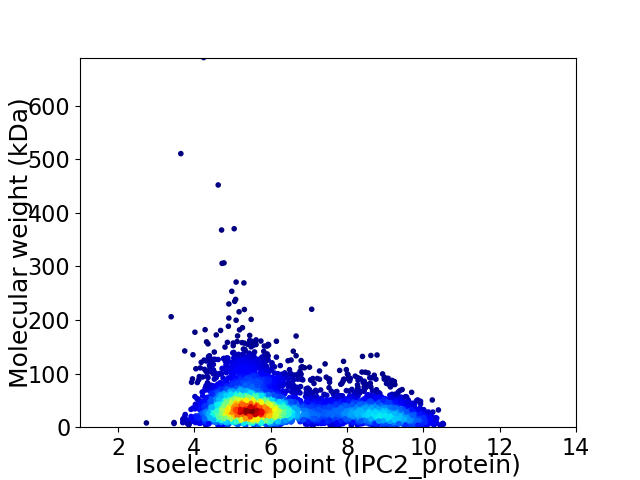
Virtual 2D-PAGE plot for 5672 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9RI04|A0A1G9RI04_9BACT PAS domain S-box-containing protein OS=Catalinimonas alkaloidigena OX=1075417 GN=SAMN05421823_111167 PE=4 SV=1
MM1 pKa = 7.1LTQVRR6 pKa = 11.84SRR8 pKa = 11.84LGAFLVLAGLSSVSLAQTPVFINEE32 pKa = 3.39IHH34 pKa = 6.5YY35 pKa = 11.13DD36 pKa = 3.61NVGTDD41 pKa = 2.96TGEE44 pKa = 4.47AIEE47 pKa = 4.25IAGPAGTDD55 pKa = 3.26LTGWSLVLYY64 pKa = 10.05NGSNNQPYY72 pKa = 7.17TTSALSGTLADD83 pKa = 4.38AGSGYY88 pKa = 10.68GFAVVTYY95 pKa = 8.29PSNGIQNGDD104 pKa = 3.67PDD106 pKa = 4.27GVALVNSAGAVIQFLSYY123 pKa = 10.95DD124 pKa = 3.58GTMTAATGPAQGLTSIDD141 pKa = 3.2IGVGEE146 pKa = 4.75SNATTTGFSLQLTGSGSSYY165 pKa = 11.55EE166 pKa = 4.12DD167 pKa = 3.52FTWADD172 pKa = 3.17AQANTFGAVNAGQTFGGGPVGPADD196 pKa = 3.42PVINEE201 pKa = 4.36FSASTTGTDD210 pKa = 3.05VEE212 pKa = 4.58YY213 pKa = 10.7IEE215 pKa = 5.84ILGDD219 pKa = 3.63PNADD223 pKa = 3.56YY224 pKa = 11.65SDD226 pKa = 3.77LTVLEE231 pKa = 4.59IEE233 pKa = 5.01GDD235 pKa = 3.73TPAQGTIDD243 pKa = 4.31GIFPLGTTDD252 pKa = 3.23ANGYY256 pKa = 9.63FLLNLPANEE265 pKa = 4.74LEE267 pKa = 4.38NGTLTLLLVRR277 pKa = 11.84NFADD281 pKa = 3.54TLGADD286 pKa = 4.42LDD288 pKa = 4.17ADD290 pKa = 3.8NDD292 pKa = 4.07GVLDD296 pKa = 3.71VTPWTDD302 pKa = 3.43LVDD305 pKa = 5.45GIAVNDD311 pKa = 4.45GGAGDD316 pKa = 3.47QTYY319 pKa = 10.86AVTLNVGYY327 pKa = 10.37DD328 pKa = 3.74GLSFAPGGASRR339 pKa = 11.84LPNGTDD345 pKa = 3.3TDD347 pKa = 3.92QTSDD351 pKa = 2.61WTRR354 pKa = 11.84NDD356 pKa = 2.99FDD358 pKa = 4.27LAGIDD363 pKa = 4.08GYY365 pKa = 10.93EE366 pKa = 4.21GSPVPGEE373 pKa = 4.4AYY375 pKa = 7.71NTPGAPNQAVAAPLPAIVINEE396 pKa = 4.27LDD398 pKa = 3.54VDD400 pKa = 4.23QDD402 pKa = 3.58GTDD405 pKa = 2.96AAEE408 pKa = 4.79FIEE411 pKa = 5.69LYY413 pKa = 10.76DD414 pKa = 3.88GGVGNTALNGLVLVLFNGNGDD435 pKa = 3.63EE436 pKa = 4.48AYY438 pKa = 10.45GAIALDD444 pKa = 4.92GYY446 pKa = 7.62TTDD449 pKa = 3.24AEE451 pKa = 5.33GYY453 pKa = 9.72FVVGSANVANVDD465 pKa = 3.79LVAFTTNGLQNGADD479 pKa = 3.56AVALYY484 pKa = 10.44QGDD487 pKa = 3.59AAAITTVTTDD497 pKa = 3.26GLIDD501 pKa = 3.54AVVYY505 pKa = 7.59GTSDD509 pKa = 3.47ADD511 pKa = 4.61DD512 pKa = 4.36EE513 pKa = 4.77GLLALLNAGQPQADD527 pKa = 3.85EE528 pKa = 4.27NANGQAISEE537 pKa = 4.31SLQRR541 pKa = 11.84LPNGSGGQRR550 pKa = 11.84NTDD553 pKa = 3.34TFGAAAPTPGTEE565 pKa = 3.95NGGDD569 pKa = 3.7GGPVEE574 pKa = 4.56PQPGLITIAEE584 pKa = 4.19ARR586 pKa = 11.84NAADD590 pKa = 3.94DD591 pKa = 4.09AVVTIQGILTVADD604 pKa = 3.96EE605 pKa = 4.9FGGPAYY611 pKa = 9.82IQDD614 pKa = 3.97STGGIAVFDD623 pKa = 4.16AALHH627 pKa = 5.97GGTYY631 pKa = 10.24QIGDD635 pKa = 3.81EE636 pKa = 4.4VKK638 pKa = 10.02LTGALDD644 pKa = 3.73TYY646 pKa = 10.97NGLRR650 pKa = 11.84EE651 pKa = 4.46VVSVTHH657 pKa = 7.15DD658 pKa = 3.67SLVSTGNSVTPAAITVAQLAAYY680 pKa = 7.92EE681 pKa = 4.37GQLVTLQNVSFPNPGDD697 pKa = 3.96LFWPNSNFVITDD709 pKa = 3.31ATGSGEE715 pKa = 4.08LRR717 pKa = 11.84IDD719 pKa = 3.76ADD721 pKa = 3.81VEE723 pKa = 4.57GIAGKK728 pKa = 9.59AQPEE732 pKa = 4.47TCEE735 pKa = 3.84VTGVIGQFNGTPQLLPRR752 pKa = 11.84LAGDD756 pKa = 4.36LPCAEE761 pKa = 5.17DD762 pKa = 4.19FVPPTGDD769 pKa = 2.96TDD771 pKa = 4.29LPLAQTLDD779 pKa = 3.58VVTWNVEE786 pKa = 3.95WFGSPSQGPTNEE798 pKa = 3.91TLQADD803 pKa = 3.87NVKK806 pKa = 10.35RR807 pKa = 11.84VLDD810 pKa = 3.8SLDD813 pKa = 3.49ADD815 pKa = 3.98VYY817 pKa = 11.04ALEE820 pKa = 5.14EE821 pKa = 4.28ISDD824 pKa = 4.05SALLQQVVDD833 pKa = 3.98QLPGYY838 pKa = 10.36ALAIQTGYY846 pKa = 10.84VSYY849 pKa = 10.25PPNEE853 pKa = 3.74PRR855 pKa = 11.84EE856 pKa = 4.19SQKK859 pKa = 11.01LAFIYY864 pKa = 9.12KK865 pKa = 8.65TEE867 pKa = 4.2TVSPVRR873 pKa = 11.84TQPLLDD879 pKa = 3.63WQHH882 pKa = 6.54PYY884 pKa = 11.41YY885 pKa = 10.97NGGDD889 pKa = 3.58DD890 pKa = 5.6SYY892 pKa = 11.32LTDD895 pKa = 3.73YY896 pKa = 10.52PADD899 pKa = 3.53DD900 pKa = 3.54RR901 pKa = 11.84TRR903 pKa = 11.84FFASGRR909 pKa = 11.84LPFLMVADD917 pKa = 3.87VTIDD921 pKa = 3.19GSTEE925 pKa = 3.64RR926 pKa = 11.84IYY928 pKa = 10.84FVGLHH933 pKa = 5.61ARR935 pKa = 11.84ANGSDD940 pKa = 3.36AQLTYY945 pKa = 11.61DD946 pKa = 3.27MRR948 pKa = 11.84KK949 pKa = 9.82YY950 pKa = 10.64DD951 pKa = 3.81VEE953 pKa = 4.01RR954 pKa = 11.84LKK956 pKa = 11.19AVLDD960 pKa = 3.82TLGGGTANVMLLGDD974 pKa = 3.99YY975 pKa = 11.01NDD977 pKa = 5.26DD978 pKa = 3.4VDD980 pKa = 3.95EE981 pKa = 4.46TVANISSTVTSYY993 pKa = 11.4AAYY996 pKa = 10.41VQDD999 pKa = 5.28DD1000 pKa = 4.33SLNPGADD1007 pKa = 3.14GNYY1010 pKa = 9.36YY1011 pKa = 10.84AVLTSSLSDD1020 pKa = 2.94AGFRR1024 pKa = 11.84TYY1026 pKa = 11.05VSYY1029 pKa = 11.42EE1030 pKa = 3.73NVIDD1034 pKa = 5.1HH1035 pKa = 6.19ITVTDD1040 pKa = 4.01EE1041 pKa = 4.02LTDD1044 pKa = 3.56NYY1046 pKa = 9.67IANSATVHH1054 pKa = 5.61YY1055 pKa = 9.88EE1056 pKa = 3.75VFAGNSVIPNYY1067 pKa = 10.99ASTTSDD1073 pKa = 3.04HH1074 pKa = 6.34MPVSVRR1080 pKa = 11.84LVLEE1084 pKa = 4.04KK1085 pKa = 10.95APALTLGSCVYY1096 pKa = 9.72TVEE1099 pKa = 4.78DD1100 pKa = 4.35FPGAATITALDD1111 pKa = 3.9SVTYY1115 pKa = 9.8WDD1117 pKa = 4.22FFQRR1121 pKa = 11.84RR1122 pKa = 11.84GSYY1125 pKa = 8.88TVGVATGVRR1134 pKa = 11.84TDD1136 pKa = 3.5GQAGAWEE1143 pKa = 4.03IHH1145 pKa = 6.38NDD1147 pKa = 3.61CSIKK1151 pKa = 10.19PLRR1154 pKa = 11.84KK1155 pKa = 9.71SGSKK1159 pKa = 10.49QNTSLLPDD1167 pKa = 3.23VLGAKK1172 pKa = 10.12RR1173 pKa = 11.84KK1174 pKa = 10.21DD1175 pKa = 3.01GWHH1178 pKa = 5.87YY1179 pKa = 10.65EE1180 pKa = 4.08PYY1182 pKa = 10.41AISDD1186 pKa = 3.56DD1187 pKa = 4.06GEE1189 pKa = 4.47YY1190 pKa = 10.59ILANAINTDD1199 pKa = 3.33GYY1201 pKa = 9.81TIGRR1205 pKa = 11.84WRR1207 pKa = 11.84GVAPGATLPVKK1218 pKa = 10.38FRR1220 pKa = 11.84IGGPLLGRR1228 pKa = 11.84VFILLGTVDD1237 pKa = 4.74CDD1239 pKa = 3.75DD1240 pKa = 5.81LAMEE1244 pKa = 4.39WTDD1247 pKa = 3.88GRR1249 pKa = 11.84YY1250 pKa = 9.95LLTGCDD1256 pKa = 3.48ASEE1259 pKa = 4.03EE1260 pKa = 4.42VPTRR1264 pKa = 11.84VIASVPVQTAQTHH1277 pKa = 6.83LYY1279 pKa = 8.73PNPARR1284 pKa = 11.84QSITVQFAEE1293 pKa = 4.48SLQSNATLRR1302 pKa = 11.84VVDD1305 pKa = 3.89ALGRR1309 pKa = 11.84LYY1311 pKa = 10.98AEE1313 pKa = 4.19RR1314 pKa = 11.84TVEE1317 pKa = 3.95AGQVEE1322 pKa = 4.57YY1323 pKa = 10.99QEE1325 pKa = 4.35QLSNLPSGIYY1335 pKa = 10.29FMQVQTAEE1343 pKa = 4.42GVQNLRR1349 pKa = 11.84FVKK1352 pKa = 10.31QQ1353 pKa = 3.15
MM1 pKa = 7.1LTQVRR6 pKa = 11.84SRR8 pKa = 11.84LGAFLVLAGLSSVSLAQTPVFINEE32 pKa = 3.39IHH34 pKa = 6.5YY35 pKa = 11.13DD36 pKa = 3.61NVGTDD41 pKa = 2.96TGEE44 pKa = 4.47AIEE47 pKa = 4.25IAGPAGTDD55 pKa = 3.26LTGWSLVLYY64 pKa = 10.05NGSNNQPYY72 pKa = 7.17TTSALSGTLADD83 pKa = 4.38AGSGYY88 pKa = 10.68GFAVVTYY95 pKa = 8.29PSNGIQNGDD104 pKa = 3.67PDD106 pKa = 4.27GVALVNSAGAVIQFLSYY123 pKa = 10.95DD124 pKa = 3.58GTMTAATGPAQGLTSIDD141 pKa = 3.2IGVGEE146 pKa = 4.75SNATTTGFSLQLTGSGSSYY165 pKa = 11.55EE166 pKa = 4.12DD167 pKa = 3.52FTWADD172 pKa = 3.17AQANTFGAVNAGQTFGGGPVGPADD196 pKa = 3.42PVINEE201 pKa = 4.36FSASTTGTDD210 pKa = 3.05VEE212 pKa = 4.58YY213 pKa = 10.7IEE215 pKa = 5.84ILGDD219 pKa = 3.63PNADD223 pKa = 3.56YY224 pKa = 11.65SDD226 pKa = 3.77LTVLEE231 pKa = 4.59IEE233 pKa = 5.01GDD235 pKa = 3.73TPAQGTIDD243 pKa = 4.31GIFPLGTTDD252 pKa = 3.23ANGYY256 pKa = 9.63FLLNLPANEE265 pKa = 4.74LEE267 pKa = 4.38NGTLTLLLVRR277 pKa = 11.84NFADD281 pKa = 3.54TLGADD286 pKa = 4.42LDD288 pKa = 4.17ADD290 pKa = 3.8NDD292 pKa = 4.07GVLDD296 pKa = 3.71VTPWTDD302 pKa = 3.43LVDD305 pKa = 5.45GIAVNDD311 pKa = 4.45GGAGDD316 pKa = 3.47QTYY319 pKa = 10.86AVTLNVGYY327 pKa = 10.37DD328 pKa = 3.74GLSFAPGGASRR339 pKa = 11.84LPNGTDD345 pKa = 3.3TDD347 pKa = 3.92QTSDD351 pKa = 2.61WTRR354 pKa = 11.84NDD356 pKa = 2.99FDD358 pKa = 4.27LAGIDD363 pKa = 4.08GYY365 pKa = 10.93EE366 pKa = 4.21GSPVPGEE373 pKa = 4.4AYY375 pKa = 7.71NTPGAPNQAVAAPLPAIVINEE396 pKa = 4.27LDD398 pKa = 3.54VDD400 pKa = 4.23QDD402 pKa = 3.58GTDD405 pKa = 2.96AAEE408 pKa = 4.79FIEE411 pKa = 5.69LYY413 pKa = 10.76DD414 pKa = 3.88GGVGNTALNGLVLVLFNGNGDD435 pKa = 3.63EE436 pKa = 4.48AYY438 pKa = 10.45GAIALDD444 pKa = 4.92GYY446 pKa = 7.62TTDD449 pKa = 3.24AEE451 pKa = 5.33GYY453 pKa = 9.72FVVGSANVANVDD465 pKa = 3.79LVAFTTNGLQNGADD479 pKa = 3.56AVALYY484 pKa = 10.44QGDD487 pKa = 3.59AAAITTVTTDD497 pKa = 3.26GLIDD501 pKa = 3.54AVVYY505 pKa = 7.59GTSDD509 pKa = 3.47ADD511 pKa = 4.61DD512 pKa = 4.36EE513 pKa = 4.77GLLALLNAGQPQADD527 pKa = 3.85EE528 pKa = 4.27NANGQAISEE537 pKa = 4.31SLQRR541 pKa = 11.84LPNGSGGQRR550 pKa = 11.84NTDD553 pKa = 3.34TFGAAAPTPGTEE565 pKa = 3.95NGGDD569 pKa = 3.7GGPVEE574 pKa = 4.56PQPGLITIAEE584 pKa = 4.19ARR586 pKa = 11.84NAADD590 pKa = 3.94DD591 pKa = 4.09AVVTIQGILTVADD604 pKa = 3.96EE605 pKa = 4.9FGGPAYY611 pKa = 9.82IQDD614 pKa = 3.97STGGIAVFDD623 pKa = 4.16AALHH627 pKa = 5.97GGTYY631 pKa = 10.24QIGDD635 pKa = 3.81EE636 pKa = 4.4VKK638 pKa = 10.02LTGALDD644 pKa = 3.73TYY646 pKa = 10.97NGLRR650 pKa = 11.84EE651 pKa = 4.46VVSVTHH657 pKa = 7.15DD658 pKa = 3.67SLVSTGNSVTPAAITVAQLAAYY680 pKa = 7.92EE681 pKa = 4.37GQLVTLQNVSFPNPGDD697 pKa = 3.96LFWPNSNFVITDD709 pKa = 3.31ATGSGEE715 pKa = 4.08LRR717 pKa = 11.84IDD719 pKa = 3.76ADD721 pKa = 3.81VEE723 pKa = 4.57GIAGKK728 pKa = 9.59AQPEE732 pKa = 4.47TCEE735 pKa = 3.84VTGVIGQFNGTPQLLPRR752 pKa = 11.84LAGDD756 pKa = 4.36LPCAEE761 pKa = 5.17DD762 pKa = 4.19FVPPTGDD769 pKa = 2.96TDD771 pKa = 4.29LPLAQTLDD779 pKa = 3.58VVTWNVEE786 pKa = 3.95WFGSPSQGPTNEE798 pKa = 3.91TLQADD803 pKa = 3.87NVKK806 pKa = 10.35RR807 pKa = 11.84VLDD810 pKa = 3.8SLDD813 pKa = 3.49ADD815 pKa = 3.98VYY817 pKa = 11.04ALEE820 pKa = 5.14EE821 pKa = 4.28ISDD824 pKa = 4.05SALLQQVVDD833 pKa = 3.98QLPGYY838 pKa = 10.36ALAIQTGYY846 pKa = 10.84VSYY849 pKa = 10.25PPNEE853 pKa = 3.74PRR855 pKa = 11.84EE856 pKa = 4.19SQKK859 pKa = 11.01LAFIYY864 pKa = 9.12KK865 pKa = 8.65TEE867 pKa = 4.2TVSPVRR873 pKa = 11.84TQPLLDD879 pKa = 3.63WQHH882 pKa = 6.54PYY884 pKa = 11.41YY885 pKa = 10.97NGGDD889 pKa = 3.58DD890 pKa = 5.6SYY892 pKa = 11.32LTDD895 pKa = 3.73YY896 pKa = 10.52PADD899 pKa = 3.53DD900 pKa = 3.54RR901 pKa = 11.84TRR903 pKa = 11.84FFASGRR909 pKa = 11.84LPFLMVADD917 pKa = 3.87VTIDD921 pKa = 3.19GSTEE925 pKa = 3.64RR926 pKa = 11.84IYY928 pKa = 10.84FVGLHH933 pKa = 5.61ARR935 pKa = 11.84ANGSDD940 pKa = 3.36AQLTYY945 pKa = 11.61DD946 pKa = 3.27MRR948 pKa = 11.84KK949 pKa = 9.82YY950 pKa = 10.64DD951 pKa = 3.81VEE953 pKa = 4.01RR954 pKa = 11.84LKK956 pKa = 11.19AVLDD960 pKa = 3.82TLGGGTANVMLLGDD974 pKa = 3.99YY975 pKa = 11.01NDD977 pKa = 5.26DD978 pKa = 3.4VDD980 pKa = 3.95EE981 pKa = 4.46TVANISSTVTSYY993 pKa = 11.4AAYY996 pKa = 10.41VQDD999 pKa = 5.28DD1000 pKa = 4.33SLNPGADD1007 pKa = 3.14GNYY1010 pKa = 9.36YY1011 pKa = 10.84AVLTSSLSDD1020 pKa = 2.94AGFRR1024 pKa = 11.84TYY1026 pKa = 11.05VSYY1029 pKa = 11.42EE1030 pKa = 3.73NVIDD1034 pKa = 5.1HH1035 pKa = 6.19ITVTDD1040 pKa = 4.01EE1041 pKa = 4.02LTDD1044 pKa = 3.56NYY1046 pKa = 9.67IANSATVHH1054 pKa = 5.61YY1055 pKa = 9.88EE1056 pKa = 3.75VFAGNSVIPNYY1067 pKa = 10.99ASTTSDD1073 pKa = 3.04HH1074 pKa = 6.34MPVSVRR1080 pKa = 11.84LVLEE1084 pKa = 4.04KK1085 pKa = 10.95APALTLGSCVYY1096 pKa = 9.72TVEE1099 pKa = 4.78DD1100 pKa = 4.35FPGAATITALDD1111 pKa = 3.9SVTYY1115 pKa = 9.8WDD1117 pKa = 4.22FFQRR1121 pKa = 11.84RR1122 pKa = 11.84GSYY1125 pKa = 8.88TVGVATGVRR1134 pKa = 11.84TDD1136 pKa = 3.5GQAGAWEE1143 pKa = 4.03IHH1145 pKa = 6.38NDD1147 pKa = 3.61CSIKK1151 pKa = 10.19PLRR1154 pKa = 11.84KK1155 pKa = 9.71SGSKK1159 pKa = 10.49QNTSLLPDD1167 pKa = 3.23VLGAKK1172 pKa = 10.12RR1173 pKa = 11.84KK1174 pKa = 10.21DD1175 pKa = 3.01GWHH1178 pKa = 5.87YY1179 pKa = 10.65EE1180 pKa = 4.08PYY1182 pKa = 10.41AISDD1186 pKa = 3.56DD1187 pKa = 4.06GEE1189 pKa = 4.47YY1190 pKa = 10.59ILANAINTDD1199 pKa = 3.33GYY1201 pKa = 9.81TIGRR1205 pKa = 11.84WRR1207 pKa = 11.84GVAPGATLPVKK1218 pKa = 10.38FRR1220 pKa = 11.84IGGPLLGRR1228 pKa = 11.84VFILLGTVDD1237 pKa = 4.74CDD1239 pKa = 3.75DD1240 pKa = 5.81LAMEE1244 pKa = 4.39WTDD1247 pKa = 3.88GRR1249 pKa = 11.84YY1250 pKa = 9.95LLTGCDD1256 pKa = 3.48ASEE1259 pKa = 4.03EE1260 pKa = 4.42VPTRR1264 pKa = 11.84VIASVPVQTAQTHH1277 pKa = 6.83LYY1279 pKa = 8.73PNPARR1284 pKa = 11.84QSITVQFAEE1293 pKa = 4.48SLQSNATLRR1302 pKa = 11.84VVDD1305 pKa = 3.89ALGRR1309 pKa = 11.84LYY1311 pKa = 10.98AEE1313 pKa = 4.19RR1314 pKa = 11.84TVEE1317 pKa = 3.95AGQVEE1322 pKa = 4.57YY1323 pKa = 10.99QEE1325 pKa = 4.35QLSNLPSGIYY1335 pKa = 10.29FMQVQTAEE1343 pKa = 4.42GVQNLRR1349 pKa = 11.84FVKK1352 pKa = 10.31QQ1353 pKa = 3.15
Molecular weight: 142.32 kDa
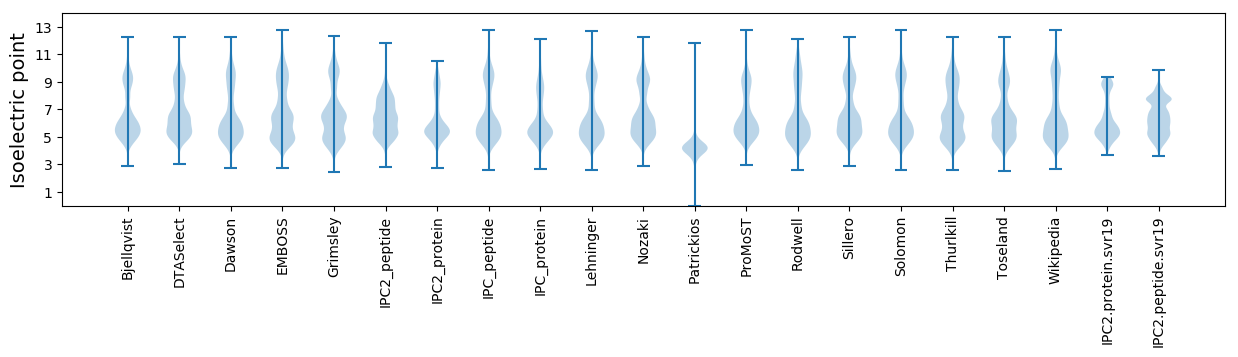
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G8ZXG0|A0A1G8ZXG0_9BACT DNA-binding response regulator OmpR family contains REC and winged-helix (WHTH) domain OS=Catalinimonas alkaloidigena OX=1075417 GN=SAMN05421823_102117 PE=4 SV=1
MM1 pKa = 7.5RR2 pKa = 11.84QPVDD6 pKa = 2.91HH7 pKa = 6.34STIANVLSSRR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.91ATTHH23 pKa = 5.88AQNPPPKK30 pKa = 9.51EE31 pKa = 3.83RR32 pKa = 11.84RR33 pKa = 11.84LHH35 pKa = 5.8YY36 pKa = 10.3SLHH39 pKa = 5.63TAVTPTLRR47 pKa = 11.84TLEE50 pKa = 4.48HH51 pKa = 6.87DD52 pKa = 5.04HH53 pKa = 6.63ICLAYY58 pKa = 9.84RR59 pKa = 11.84VWGTGTHH66 pKa = 6.58PMLAFHH72 pKa = 7.21GFGQTGAHH80 pKa = 6.54FAALQPAWEE89 pKa = 4.29HH90 pKa = 5.17TFRR93 pKa = 11.84VYY95 pKa = 11.15AFDD98 pKa = 4.79LFCHH102 pKa = 6.68GDD104 pKa = 3.7SAWPHH109 pKa = 4.5GTMPLNFATWTTLMQRR125 pKa = 11.84FCQQEE130 pKa = 4.25KK131 pKa = 10.23IDD133 pKa = 4.1MFSAAGFSLGSRR145 pKa = 11.84LALATATAFPQQLTSLTLVAPDD167 pKa = 4.61SPFLSPWYY175 pKa = 8.99RR176 pKa = 11.84LAMAWPRR183 pKa = 11.84LFRR186 pKa = 11.84WSLSHH191 pKa = 7.29SDD193 pKa = 3.65VWLSWITRR201 pKa = 11.84LEE203 pKa = 4.11QTGVLAPAAARR214 pKa = 11.84LALRR218 pKa = 11.84SLRR221 pKa = 11.84QPAHH225 pKa = 7.07RR226 pKa = 11.84ARR228 pKa = 11.84MACAWPVFSALRR240 pKa = 11.84FSPVSLSQLFEE251 pKa = 3.86RR252 pKa = 11.84HH253 pKa = 5.49AVSVQLFLGKK263 pKa = 9.86NDD265 pKa = 4.86PIAAEE270 pKa = 4.11RR271 pKa = 11.84KK272 pKa = 8.8LIAWVGPMTRR282 pKa = 11.84GKK284 pKa = 8.92VTLLDD289 pKa = 3.87VPHH292 pKa = 6.75EE293 pKa = 4.1RR294 pKa = 11.84LLAVVAGHH302 pKa = 7.09WPQNN306 pKa = 3.52
MM1 pKa = 7.5RR2 pKa = 11.84QPVDD6 pKa = 2.91HH7 pKa = 6.34STIANVLSSRR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.91ATTHH23 pKa = 5.88AQNPPPKK30 pKa = 9.51EE31 pKa = 3.83RR32 pKa = 11.84RR33 pKa = 11.84LHH35 pKa = 5.8YY36 pKa = 10.3SLHH39 pKa = 5.63TAVTPTLRR47 pKa = 11.84TLEE50 pKa = 4.48HH51 pKa = 6.87DD52 pKa = 5.04HH53 pKa = 6.63ICLAYY58 pKa = 9.84RR59 pKa = 11.84VWGTGTHH66 pKa = 6.58PMLAFHH72 pKa = 7.21GFGQTGAHH80 pKa = 6.54FAALQPAWEE89 pKa = 4.29HH90 pKa = 5.17TFRR93 pKa = 11.84VYY95 pKa = 11.15AFDD98 pKa = 4.79LFCHH102 pKa = 6.68GDD104 pKa = 3.7SAWPHH109 pKa = 4.5GTMPLNFATWTTLMQRR125 pKa = 11.84FCQQEE130 pKa = 4.25KK131 pKa = 10.23IDD133 pKa = 4.1MFSAAGFSLGSRR145 pKa = 11.84LALATATAFPQQLTSLTLVAPDD167 pKa = 4.61SPFLSPWYY175 pKa = 8.99RR176 pKa = 11.84LAMAWPRR183 pKa = 11.84LFRR186 pKa = 11.84WSLSHH191 pKa = 7.29SDD193 pKa = 3.65VWLSWITRR201 pKa = 11.84LEE203 pKa = 4.11QTGVLAPAAARR214 pKa = 11.84LALRR218 pKa = 11.84SLRR221 pKa = 11.84QPAHH225 pKa = 7.07RR226 pKa = 11.84ARR228 pKa = 11.84MACAWPVFSALRR240 pKa = 11.84FSPVSLSQLFEE251 pKa = 3.86RR252 pKa = 11.84HH253 pKa = 5.49AVSVQLFLGKK263 pKa = 9.86NDD265 pKa = 4.86PIAAEE270 pKa = 4.11RR271 pKa = 11.84KK272 pKa = 8.8LIAWVGPMTRR282 pKa = 11.84GKK284 pKa = 8.92VTLLDD289 pKa = 3.87VPHH292 pKa = 6.75EE293 pKa = 4.1RR294 pKa = 11.84LLAVVAGHH302 pKa = 7.09WPQNN306 pKa = 3.52
Molecular weight: 34.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2097683 |
25 |
6554 |
369.8 |
41.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.819 ± 0.033 | 0.737 ± 0.012 |
5.43 ± 0.026 | 6.082 ± 0.032 |
4.433 ± 0.022 | 7.229 ± 0.04 |
2.328 ± 0.018 | 4.879 ± 0.025 |
4.016 ± 0.035 | 10.56 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.095 ± 0.015 | 3.889 ± 0.034 |
4.782 ± 0.023 | 4.736 ± 0.027 |
5.896 ± 0.028 | 5.589 ± 0.022 |
6.097 ± 0.039 | 7.003 ± 0.028 |
1.496 ± 0.014 | 3.903 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
