
Narcissus common latent virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Quinvirinae; Carlavirus
Average proteome isoelectric point is 7.31
Get precalculated fractions of proteins
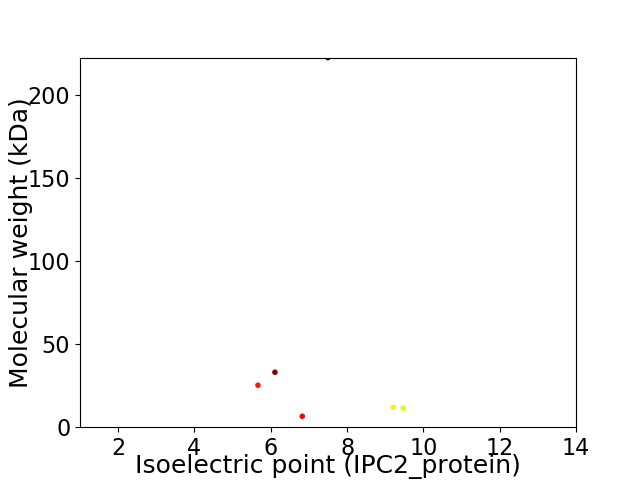
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
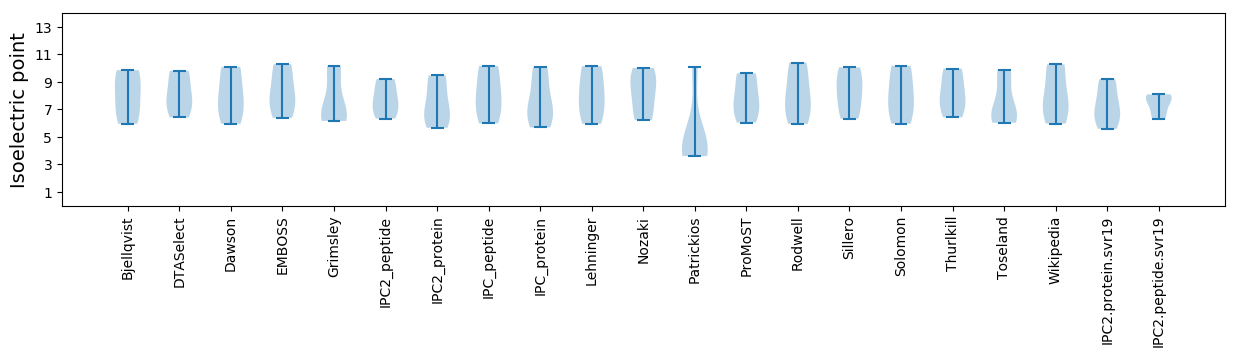
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q0VZD1|Q0VZD1_9VIRU Helicase OS=Narcissus common latent virus OX=160844 GN=NcLaVgp1 PE=3 SV=1
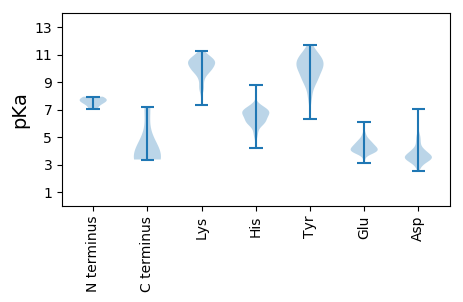
MM1 pKa = 7.82DD2 pKa = 3.99VLVNLLNKK10 pKa = 10.09YY11 pKa = 10.15GLVRR15 pKa = 11.84LSSKK19 pKa = 10.45LVLPIVVHH27 pKa = 6.02CVPGAGKK34 pKa = 8.46STLIRR39 pKa = 11.84EE40 pKa = 4.57LLACDD45 pKa = 3.72SRR47 pKa = 11.84FVAYY51 pKa = 9.28TGGLGDD57 pKa = 4.14PAHH60 pKa = 6.68ITGKK64 pKa = 9.5WIQRR68 pKa = 11.84WQGDD72 pKa = 3.38YY73 pKa = 11.42DD74 pKa = 3.6RR75 pKa = 11.84TKK77 pKa = 11.25NLVLDD82 pKa = 4.97EE83 pKa = 4.3YY84 pKa = 10.94TLIEE88 pKa = 4.5DD89 pKa = 3.77VPGAFALFGDD99 pKa = 5.27PIQVNTTSVKK109 pKa = 9.87PADD112 pKa = 4.65FICYY116 pKa = 6.72TSRR119 pKa = 11.84RR120 pKa = 11.84FGSATSTLLKK130 pKa = 10.74ALGWPVEE137 pKa = 3.97ASGNDD142 pKa = 3.47LVQIHH147 pKa = 7.1HH148 pKa = 7.14IYY150 pKa = 10.48SFEE153 pKa = 3.9PTGVVIYY160 pKa = 9.99FEE162 pKa = 4.76EE163 pKa = 4.96EE164 pKa = 3.56IGCLLRR170 pKa = 11.84EE171 pKa = 4.25HH172 pKa = 6.73CVAALGLEE180 pKa = 4.42EE181 pKa = 4.7IRR183 pKa = 11.84GKK185 pKa = 8.75TFDD188 pKa = 3.18TVTFVTSEE196 pKa = 3.87NSPLISRR203 pKa = 11.84EE204 pKa = 3.64AAYY207 pKa = 10.35QCLTRR212 pKa = 11.84HH213 pKa = 6.02RR214 pKa = 11.84LALHH218 pKa = 6.31ILCPNATYY226 pKa = 7.8TAPP229 pKa = 3.55
MM1 pKa = 7.82DD2 pKa = 3.99VLVNLLNKK10 pKa = 10.09YY11 pKa = 10.15GLVRR15 pKa = 11.84LSSKK19 pKa = 10.45LVLPIVVHH27 pKa = 6.02CVPGAGKK34 pKa = 8.46STLIRR39 pKa = 11.84EE40 pKa = 4.57LLACDD45 pKa = 3.72SRR47 pKa = 11.84FVAYY51 pKa = 9.28TGGLGDD57 pKa = 4.14PAHH60 pKa = 6.68ITGKK64 pKa = 9.5WIQRR68 pKa = 11.84WQGDD72 pKa = 3.38YY73 pKa = 11.42DD74 pKa = 3.6RR75 pKa = 11.84TKK77 pKa = 11.25NLVLDD82 pKa = 4.97EE83 pKa = 4.3YY84 pKa = 10.94TLIEE88 pKa = 4.5DD89 pKa = 3.77VPGAFALFGDD99 pKa = 5.27PIQVNTTSVKK109 pKa = 9.87PADD112 pKa = 4.65FICYY116 pKa = 6.72TSRR119 pKa = 11.84RR120 pKa = 11.84FGSATSTLLKK130 pKa = 10.74ALGWPVEE137 pKa = 3.97ASGNDD142 pKa = 3.47LVQIHH147 pKa = 7.1HH148 pKa = 7.14IYY150 pKa = 10.48SFEE153 pKa = 3.9PTGVVIYY160 pKa = 9.99FEE162 pKa = 4.76EE163 pKa = 4.96EE164 pKa = 3.56IGCLLRR170 pKa = 11.84EE171 pKa = 4.25HH172 pKa = 6.73CVAALGLEE180 pKa = 4.42EE181 pKa = 4.7IRR183 pKa = 11.84GKK185 pKa = 8.75TFDD188 pKa = 3.18TVTFVTSEE196 pKa = 3.87NSPLISRR203 pKa = 11.84EE204 pKa = 3.64AAYY207 pKa = 10.35QCLTRR212 pKa = 11.84HH213 pKa = 6.02RR214 pKa = 11.84LALHH218 pKa = 6.31ILCPNATYY226 pKa = 7.8TAPP229 pKa = 3.55
Molecular weight: 25.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0VZD0|Q0VZD0_9VIRU Triple Gene Block protein 1 OS=Narcissus common latent virus OX=160844 GN=NcLaVgp2 PE=4 SV=1
MM1 pKa = 7.93PLTPPRR7 pKa = 11.84DD8 pKa = 3.58FTKK11 pKa = 10.71VYY13 pKa = 9.9IAASVGATLALVTWLLTKK31 pKa = 9.83NTLPAVGDD39 pKa = 3.64RR40 pKa = 11.84DD41 pKa = 3.62HH42 pKa = 6.88NLPHH46 pKa = 6.8GGLYY50 pKa = 9.99RR51 pKa = 11.84DD52 pKa = 3.49GTKK55 pKa = 10.23AIKK58 pKa = 10.5YY59 pKa = 7.91NSPCKK64 pKa = 10.18LNSIEE69 pKa = 4.06GHH71 pKa = 6.24SRR73 pKa = 11.84GIWNQPWALVFILSVLIIISNRR95 pKa = 11.84LDD97 pKa = 3.18SRR99 pKa = 11.84VCRR102 pKa = 11.84RR103 pKa = 11.84CGTTHH108 pKa = 7.2
MM1 pKa = 7.93PLTPPRR7 pKa = 11.84DD8 pKa = 3.58FTKK11 pKa = 10.71VYY13 pKa = 9.9IAASVGATLALVTWLLTKK31 pKa = 9.83NTLPAVGDD39 pKa = 3.64RR40 pKa = 11.84DD41 pKa = 3.62HH42 pKa = 6.88NLPHH46 pKa = 6.8GGLYY50 pKa = 9.99RR51 pKa = 11.84DD52 pKa = 3.49GTKK55 pKa = 10.23AIKK58 pKa = 10.5YY59 pKa = 7.91NSPCKK64 pKa = 10.18LNSIEE69 pKa = 4.06GHH71 pKa = 6.24SRR73 pKa = 11.84GIWNQPWALVFILSVLIIISNRR95 pKa = 11.84LDD97 pKa = 3.18SRR99 pKa = 11.84VCRR102 pKa = 11.84RR103 pKa = 11.84CGTTHH108 pKa = 7.2
Molecular weight: 11.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2767 |
61 |
1966 |
461.2 |
51.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.987 ± 0.768 | 2.783 ± 0.421 |
4.445 ± 0.593 | 5.891 ± 0.741 |
5.204 ± 0.825 | 5.819 ± 0.611 |
2.927 ± 0.488 | 4.843 ± 0.445 |
5.313 ± 0.838 | 10.481 ± 0.782 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.879 ± 0.287 | 4.192 ± 0.473 |
4.662 ± 0.341 | 3.144 ± 0.425 |
6.108 ± 0.702 | 7.264 ± 0.28 |
5.891 ± 0.789 | 6.397 ± 0.738 |
1.265 ± 0.15 | 3.506 ± 0.487 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
