
Microviridae phi-CA82
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
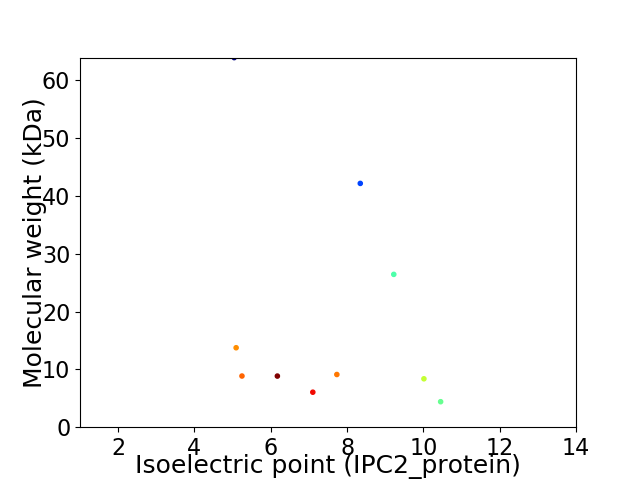
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8RW83|F8RW83_9VIRU Structural protein VP2 OS=Microviridae phi-CA82 OX=913970 GN=VP2 PE=4 SV=1
MM1 pKa = 7.61HH2 pKa = 7.08MRR4 pKa = 11.84PGFNAGLCIPIYY16 pKa = 10.32IEE18 pKa = 4.75EE19 pKa = 4.49ILPNDD24 pKa = 3.84TVEE27 pKa = 4.46MNVNMALRR35 pKa = 11.84MQTLATVPYY44 pKa = 10.6DD45 pKa = 4.12NLDD48 pKa = 3.6LDD50 pKa = 5.57LYY52 pKa = 10.26WFYY55 pKa = 11.69APNRR59 pKa = 11.84ILWDD63 pKa = 3.28GWKK66 pKa = 9.79QFCGEE71 pKa = 4.65SKK73 pKa = 10.71SAWLDD78 pKa = 3.33KK79 pKa = 10.92TPLRR83 pKa = 11.84VPVLKK88 pKa = 10.12TSGFANPDD96 pKa = 3.6DD97 pKa = 5.34GIGTVWDD104 pKa = 4.0YY105 pKa = 11.72FGLPLIKK112 pKa = 9.85EE113 pKa = 4.54TTDD116 pKa = 2.72KK117 pKa = 10.92SKK119 pKa = 10.95RR120 pKa = 11.84IAINRR125 pKa = 11.84LPFNMYY131 pKa = 10.67AKK133 pKa = 10.01VWNDD137 pKa = 2.66WFRR140 pKa = 11.84DD141 pKa = 3.46TNYY144 pKa = 10.25QEE146 pKa = 4.34EE147 pKa = 4.07ALIDD151 pKa = 3.78YY152 pKa = 9.94GNTDD156 pKa = 2.97VTYY159 pKa = 10.76DD160 pKa = 3.53KK161 pKa = 11.23SDD163 pKa = 3.5PAKK166 pKa = 10.53GGKK169 pKa = 8.55PLSANKK175 pKa = 9.5YY176 pKa = 7.72HH177 pKa = 8.07DD178 pKa = 4.3YY179 pKa = 9.58FTTCLPSPQRR189 pKa = 11.84GEE191 pKa = 4.31SVSLLDD197 pKa = 4.6GIRR200 pKa = 11.84NSVLPVRR207 pKa = 11.84AVGTEE212 pKa = 3.9LEE214 pKa = 4.44GDD216 pKa = 3.75NEE218 pKa = 5.11LGILWGLYY226 pKa = 9.7NKK228 pKa = 10.29NNEE231 pKa = 4.09QNFLPDD237 pKa = 2.59AWKK240 pKa = 9.59WYY242 pKa = 10.34DD243 pKa = 3.21MTYY246 pKa = 10.52QADD249 pKa = 3.49NSAFLHH255 pKa = 5.49QYY257 pKa = 8.46YY258 pKa = 10.58NKK260 pKa = 10.73ASGTSTAPTDD270 pKa = 3.62KK271 pKa = 10.97ATMTPANLGVDD282 pKa = 3.39IRR284 pKa = 11.84GADD287 pKa = 3.57YY288 pKa = 11.23NSNKK292 pKa = 10.24SFVDD296 pKa = 3.39AMNLTEE302 pKa = 4.36LRR304 pKa = 11.84KK305 pKa = 10.31SIAKK309 pKa = 7.86QHH311 pKa = 5.7IAEE314 pKa = 4.14VSARR318 pKa = 11.84CGNRR322 pKa = 11.84YY323 pKa = 9.87EE324 pKa = 4.2NFLLGFFGVSAPDD337 pKa = 3.24IEE339 pKa = 4.52TGRR342 pKa = 11.84TEE344 pKa = 4.15LLGINHH350 pKa = 5.56MTLNISEE357 pKa = 4.53VVQTSASEE365 pKa = 4.17EE366 pKa = 4.25GSTPQGNISGRR377 pKa = 11.84SLTNMDD383 pKa = 3.9NPHH386 pKa = 7.03AFTKK390 pKa = 10.73SFTEE394 pKa = 4.61HH395 pKa = 5.27GWLMCIAVARR405 pKa = 11.84YY406 pKa = 7.97NHH408 pKa = 6.69SYY410 pKa = 9.83SQGIEE415 pKa = 4.28KK416 pKa = 10.26KK417 pKa = 4.3WTRR420 pKa = 11.84YY421 pKa = 9.86KK422 pKa = 11.04LEE424 pKa = 4.5DD425 pKa = 3.81YY426 pKa = 11.41YY427 pKa = 11.07MPEE430 pKa = 4.16YY431 pKa = 11.22ANMGDD436 pKa = 3.54VPVYY440 pKa = 10.44LSEE443 pKa = 5.0IYY445 pKa = 9.18ATNANAQNPDD455 pKa = 3.59NLTGSDD461 pKa = 3.39GKK463 pKa = 10.56NPQEE467 pKa = 5.29DD468 pKa = 2.65IWGYY472 pKa = 7.66QEE474 pKa = 4.68YY475 pKa = 9.78GAEE478 pKa = 4.14YY479 pKa = 9.79KK480 pKa = 10.81YY481 pKa = 11.21SPNKK485 pKa = 8.42VTGSLRR491 pKa = 11.84PDD493 pKa = 2.84IKK495 pKa = 10.99GGLSPWTWCDD505 pKa = 3.3NYY507 pKa = 11.23DD508 pKa = 4.24SEE510 pKa = 4.72PTNGADD516 pKa = 4.74WIKK519 pKa = 11.21EE520 pKa = 3.71DD521 pKa = 3.98ANNINRR527 pKa = 11.84SLSIQPTAKK536 pKa = 10.08VPQLFGNFYY545 pKa = 10.63FSNKK549 pKa = 6.41HH550 pKa = 3.74TRR552 pKa = 11.84MMPVHH557 pKa = 6.54SVPGLEE563 pKa = 4.73RR564 pKa = 11.84II565 pKa = 4.02
MM1 pKa = 7.61HH2 pKa = 7.08MRR4 pKa = 11.84PGFNAGLCIPIYY16 pKa = 10.32IEE18 pKa = 4.75EE19 pKa = 4.49ILPNDD24 pKa = 3.84TVEE27 pKa = 4.46MNVNMALRR35 pKa = 11.84MQTLATVPYY44 pKa = 10.6DD45 pKa = 4.12NLDD48 pKa = 3.6LDD50 pKa = 5.57LYY52 pKa = 10.26WFYY55 pKa = 11.69APNRR59 pKa = 11.84ILWDD63 pKa = 3.28GWKK66 pKa = 9.79QFCGEE71 pKa = 4.65SKK73 pKa = 10.71SAWLDD78 pKa = 3.33KK79 pKa = 10.92TPLRR83 pKa = 11.84VPVLKK88 pKa = 10.12TSGFANPDD96 pKa = 3.6DD97 pKa = 5.34GIGTVWDD104 pKa = 4.0YY105 pKa = 11.72FGLPLIKK112 pKa = 9.85EE113 pKa = 4.54TTDD116 pKa = 2.72KK117 pKa = 10.92SKK119 pKa = 10.95RR120 pKa = 11.84IAINRR125 pKa = 11.84LPFNMYY131 pKa = 10.67AKK133 pKa = 10.01VWNDD137 pKa = 2.66WFRR140 pKa = 11.84DD141 pKa = 3.46TNYY144 pKa = 10.25QEE146 pKa = 4.34EE147 pKa = 4.07ALIDD151 pKa = 3.78YY152 pKa = 9.94GNTDD156 pKa = 2.97VTYY159 pKa = 10.76DD160 pKa = 3.53KK161 pKa = 11.23SDD163 pKa = 3.5PAKK166 pKa = 10.53GGKK169 pKa = 8.55PLSANKK175 pKa = 9.5YY176 pKa = 7.72HH177 pKa = 8.07DD178 pKa = 4.3YY179 pKa = 9.58FTTCLPSPQRR189 pKa = 11.84GEE191 pKa = 4.31SVSLLDD197 pKa = 4.6GIRR200 pKa = 11.84NSVLPVRR207 pKa = 11.84AVGTEE212 pKa = 3.9LEE214 pKa = 4.44GDD216 pKa = 3.75NEE218 pKa = 5.11LGILWGLYY226 pKa = 9.7NKK228 pKa = 10.29NNEE231 pKa = 4.09QNFLPDD237 pKa = 2.59AWKK240 pKa = 9.59WYY242 pKa = 10.34DD243 pKa = 3.21MTYY246 pKa = 10.52QADD249 pKa = 3.49NSAFLHH255 pKa = 5.49QYY257 pKa = 8.46YY258 pKa = 10.58NKK260 pKa = 10.73ASGTSTAPTDD270 pKa = 3.62KK271 pKa = 10.97ATMTPANLGVDD282 pKa = 3.39IRR284 pKa = 11.84GADD287 pKa = 3.57YY288 pKa = 11.23NSNKK292 pKa = 10.24SFVDD296 pKa = 3.39AMNLTEE302 pKa = 4.36LRR304 pKa = 11.84KK305 pKa = 10.31SIAKK309 pKa = 7.86QHH311 pKa = 5.7IAEE314 pKa = 4.14VSARR318 pKa = 11.84CGNRR322 pKa = 11.84YY323 pKa = 9.87EE324 pKa = 4.2NFLLGFFGVSAPDD337 pKa = 3.24IEE339 pKa = 4.52TGRR342 pKa = 11.84TEE344 pKa = 4.15LLGINHH350 pKa = 5.56MTLNISEE357 pKa = 4.53VVQTSASEE365 pKa = 4.17EE366 pKa = 4.25GSTPQGNISGRR377 pKa = 11.84SLTNMDD383 pKa = 3.9NPHH386 pKa = 7.03AFTKK390 pKa = 10.73SFTEE394 pKa = 4.61HH395 pKa = 5.27GWLMCIAVARR405 pKa = 11.84YY406 pKa = 7.97NHH408 pKa = 6.69SYY410 pKa = 9.83SQGIEE415 pKa = 4.28KK416 pKa = 10.26KK417 pKa = 4.3WTRR420 pKa = 11.84YY421 pKa = 9.86KK422 pKa = 11.04LEE424 pKa = 4.5DD425 pKa = 3.81YY426 pKa = 11.41YY427 pKa = 11.07MPEE430 pKa = 4.16YY431 pKa = 11.22ANMGDD436 pKa = 3.54VPVYY440 pKa = 10.44LSEE443 pKa = 5.0IYY445 pKa = 9.18ATNANAQNPDD455 pKa = 3.59NLTGSDD461 pKa = 3.39GKK463 pKa = 10.56NPQEE467 pKa = 5.29DD468 pKa = 2.65IWGYY472 pKa = 7.66QEE474 pKa = 4.68YY475 pKa = 9.78GAEE478 pKa = 4.14YY479 pKa = 9.79KK480 pKa = 10.81YY481 pKa = 11.21SPNKK485 pKa = 8.42VTGSLRR491 pKa = 11.84PDD493 pKa = 2.84IKK495 pKa = 10.99GGLSPWTWCDD505 pKa = 3.3NYY507 pKa = 11.23DD508 pKa = 4.24SEE510 pKa = 4.72PTNGADD516 pKa = 4.74WIKK519 pKa = 11.21EE520 pKa = 3.71DD521 pKa = 3.98ANNINRR527 pKa = 11.84SLSIQPTAKK536 pKa = 10.08VPQLFGNFYY545 pKa = 10.63FSNKK549 pKa = 6.41HH550 pKa = 3.74TRR552 pKa = 11.84MMPVHH557 pKa = 6.54SVPGLEE563 pKa = 4.73RR564 pKa = 11.84II565 pKa = 4.02
Molecular weight: 63.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8RW86|F8RW86_9VIRU Uncharacterized protein p5 OS=Microviridae phi-CA82 OX=913970 GN=p5 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 10.58CNVDD6 pKa = 3.33NSTVIHH12 pKa = 5.68TLSTRR17 pKa = 11.84FPPNYY22 pKa = 9.89QHH24 pKa = 7.15GKK26 pKa = 7.96PLNHH30 pKa = 7.08KK31 pKa = 8.41GLKK34 pKa = 10.19RR35 pKa = 11.84FFTVSTLLLLLLRR48 pKa = 11.84KK49 pKa = 10.06YY50 pKa = 9.1ITIYY54 pKa = 10.2IYY56 pKa = 10.86NKK58 pKa = 9.78HH59 pKa = 5.92SRR61 pKa = 11.84QKK63 pKa = 9.05VTSDD67 pKa = 2.91NRR69 pKa = 11.84NVV71 pKa = 2.98
MM1 pKa = 7.51KK2 pKa = 10.58CNVDD6 pKa = 3.33NSTVIHH12 pKa = 5.68TLSTRR17 pKa = 11.84FPPNYY22 pKa = 9.89QHH24 pKa = 7.15GKK26 pKa = 7.96PLNHH30 pKa = 7.08KK31 pKa = 8.41GLKK34 pKa = 10.19RR35 pKa = 11.84FFTVSTLLLLLLRR48 pKa = 11.84KK49 pKa = 10.06YY50 pKa = 9.1ITIYY54 pKa = 10.2IYY56 pKa = 10.86NKK58 pKa = 9.78HH59 pKa = 5.92SRR61 pKa = 11.84QKK63 pKa = 9.05VTSDD67 pKa = 2.91NRR69 pKa = 11.84NVV71 pKa = 2.98
Molecular weight: 8.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1669 |
39 |
565 |
166.9 |
19.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.351 ± 1.457 | 1.318 ± 0.427 |
5.752 ± 0.537 | 5.812 ± 0.857 |
2.936 ± 0.265 | 5.812 ± 0.595 |
2.157 ± 0.573 | 6.171 ± 1.142 |
8.149 ± 1.238 | 6.89 ± 0.534 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.415 ± 0.296 | 7.969 ± 0.926 |
3.835 ± 0.688 | 4.434 ± 0.942 |
4.793 ± 0.499 | 6.171 ± 0.604 |
6.651 ± 0.596 | 3.835 ± 0.584 |
1.738 ± 0.54 | 5.812 ± 0.738 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
