
Hubei permutotetra-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
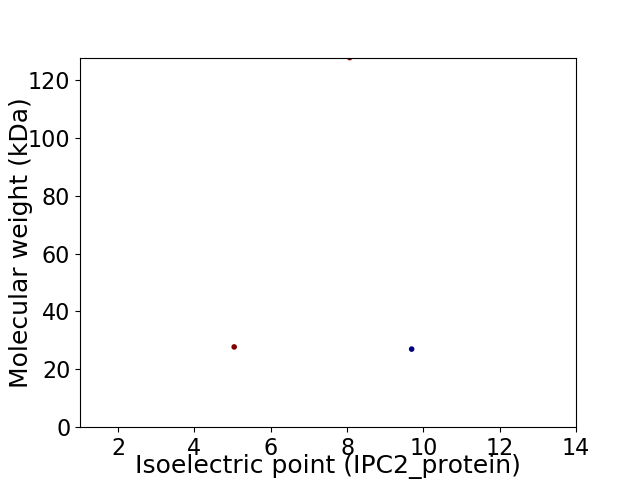
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KI36|A0A1L3KI36_9VIRU Capsid protein OS=Hubei permutotetra-like virus 9 OX=1923083 PE=3 SV=1
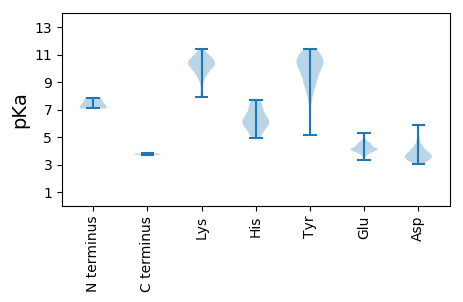
MM1 pKa = 7.11VNSEE5 pKa = 4.29GEE7 pKa = 4.19YY8 pKa = 10.09ILEE11 pKa = 4.59KK12 pKa = 10.4FQHH15 pKa = 5.85TPWNLHH21 pKa = 5.15PTPHH25 pKa = 5.68QVQGADD31 pKa = 3.68GYY33 pKa = 9.27SQYY36 pKa = 11.37VGWRR40 pKa = 11.84PSPSGPPTLFDD51 pKa = 3.64AVSLSVGNSAGFADD65 pKa = 5.64ASLQNLDD72 pKa = 3.51VEE74 pKa = 4.57EE75 pKa = 4.66PKK77 pKa = 10.71VDD79 pKa = 4.32FVFNLQPLDD88 pKa = 3.91LLQPNIPSIVRR99 pKa = 11.84LTFTLTFEE107 pKa = 4.4RR108 pKa = 11.84MNIPAEE114 pKa = 4.05EE115 pKa = 4.09NKK117 pKa = 10.4YY118 pKa = 11.15DD119 pKa = 3.57WVFDD123 pKa = 4.16LSDD126 pKa = 4.11LLDD129 pKa = 4.03HH130 pKa = 7.11VEE132 pKa = 4.13VLPTSSLVVNPHH144 pKa = 5.94LRR146 pKa = 11.84QFTKK150 pKa = 10.2TFWGFTKK157 pKa = 10.24RR158 pKa = 11.84YY159 pKa = 8.38ARR161 pKa = 11.84GYY163 pKa = 7.75IPKK166 pKa = 9.36VVVSMVSHH174 pKa = 6.67FGVEE178 pKa = 4.01AEE180 pKa = 4.12DD181 pKa = 3.8TFVGRR186 pKa = 11.84IRR188 pKa = 11.84GACEE192 pKa = 3.36WMSTSLALFTGPVVRR207 pKa = 11.84EE208 pKa = 3.94PARR211 pKa = 11.84SFAYY215 pKa = 10.01LWEE218 pKa = 4.68LNLLKK223 pKa = 10.33KK224 pKa = 10.28RR225 pKa = 11.84HH226 pKa = 5.84SRR228 pKa = 11.84PLDD231 pKa = 3.75PVDD234 pKa = 4.99DD235 pKa = 4.02SLEE238 pKa = 4.03FVLLDD243 pKa = 3.76ASS245 pKa = 3.81
MM1 pKa = 7.11VNSEE5 pKa = 4.29GEE7 pKa = 4.19YY8 pKa = 10.09ILEE11 pKa = 4.59KK12 pKa = 10.4FQHH15 pKa = 5.85TPWNLHH21 pKa = 5.15PTPHH25 pKa = 5.68QVQGADD31 pKa = 3.68GYY33 pKa = 9.27SQYY36 pKa = 11.37VGWRR40 pKa = 11.84PSPSGPPTLFDD51 pKa = 3.64AVSLSVGNSAGFADD65 pKa = 5.64ASLQNLDD72 pKa = 3.51VEE74 pKa = 4.57EE75 pKa = 4.66PKK77 pKa = 10.71VDD79 pKa = 4.32FVFNLQPLDD88 pKa = 3.91LLQPNIPSIVRR99 pKa = 11.84LTFTLTFEE107 pKa = 4.4RR108 pKa = 11.84MNIPAEE114 pKa = 4.05EE115 pKa = 4.09NKK117 pKa = 10.4YY118 pKa = 11.15DD119 pKa = 3.57WVFDD123 pKa = 4.16LSDD126 pKa = 4.11LLDD129 pKa = 4.03HH130 pKa = 7.11VEE132 pKa = 4.13VLPTSSLVVNPHH144 pKa = 5.94LRR146 pKa = 11.84QFTKK150 pKa = 10.2TFWGFTKK157 pKa = 10.24RR158 pKa = 11.84YY159 pKa = 8.38ARR161 pKa = 11.84GYY163 pKa = 7.75IPKK166 pKa = 9.36VVVSMVSHH174 pKa = 6.67FGVEE178 pKa = 4.01AEE180 pKa = 4.12DD181 pKa = 3.8TFVGRR186 pKa = 11.84IRR188 pKa = 11.84GACEE192 pKa = 3.36WMSTSLALFTGPVVRR207 pKa = 11.84EE208 pKa = 3.94PARR211 pKa = 11.84SFAYY215 pKa = 10.01LWEE218 pKa = 4.68LNLLKK223 pKa = 10.33KK224 pKa = 10.28RR225 pKa = 11.84HH226 pKa = 5.84SRR228 pKa = 11.84PLDD231 pKa = 3.75PVDD234 pKa = 4.99DD235 pKa = 4.02SLEE238 pKa = 4.03FVLLDD243 pKa = 3.76ASS245 pKa = 3.81
Molecular weight: 27.74 kDa
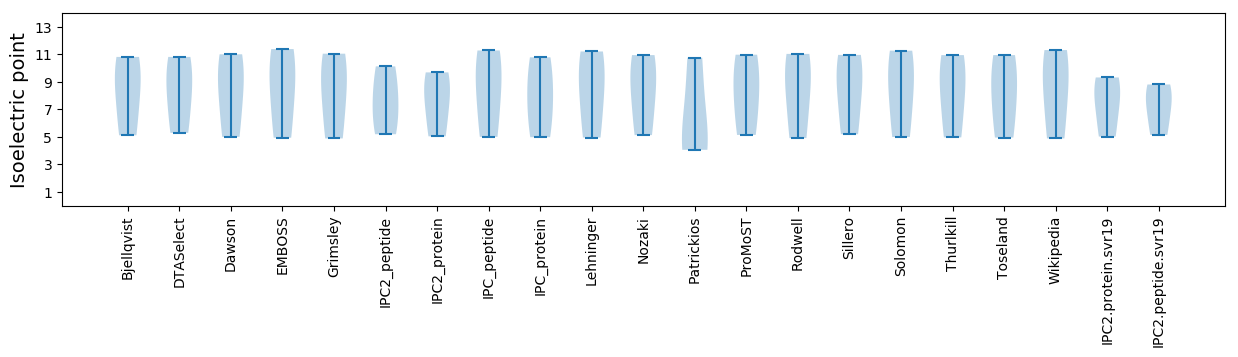
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI36|A0A1L3KI36_9VIRU Capsid protein OS=Hubei permutotetra-like virus 9 OX=1923083 PE=3 SV=1
MM1 pKa = 7.08TGRR4 pKa = 11.84RR5 pKa = 11.84ANSSSEE11 pKa = 3.76KK12 pKa = 10.28LKK14 pKa = 11.13RR15 pKa = 11.84EE16 pKa = 3.48IANLQRR22 pKa = 11.84RR23 pKa = 11.84LATSTISSNGPRR35 pKa = 11.84SGRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84GGNGNGGGGPSVPAAMGSNPRR63 pKa = 11.84PSRR66 pKa = 11.84GRR68 pKa = 11.84RR69 pKa = 11.84GGARR73 pKa = 11.84TRR75 pKa = 11.84VGNGGRR81 pKa = 11.84IVLTRR86 pKa = 11.84DD87 pKa = 3.06EE88 pKa = 4.88LLVTVATTPDD98 pKa = 3.32KK99 pKa = 11.42NEE101 pKa = 4.27SVFSKK106 pKa = 10.91DD107 pKa = 3.61LVPSPGVMPFLFRR120 pKa = 11.84LSSCYY125 pKa = 8.83QRR127 pKa = 11.84IRR129 pKa = 11.84WVRR132 pKa = 11.84ASISWRR138 pKa = 11.84PSCGTATDD146 pKa = 5.14GIISYY151 pKa = 10.33GVAFNGSEE159 pKa = 4.32SIKK162 pKa = 10.97SRR164 pKa = 11.84DD165 pKa = 3.56LVTSLTPCNDD175 pKa = 3.01HH176 pKa = 7.08PVWQSTGISPLVIPGDD192 pKa = 3.75MLMSRR197 pKa = 11.84RR198 pKa = 11.84WYY200 pKa = 8.95PLNVTGGDD208 pKa = 3.5QFDD211 pKa = 3.46KK212 pKa = 11.4GMGKK216 pKa = 9.69FCVGLTHH223 pKa = 7.65DD224 pKa = 4.25AEE226 pKa = 4.62KK227 pKa = 10.42VAKK230 pKa = 10.22SRR232 pKa = 11.84GEE234 pKa = 3.61FWISYY239 pKa = 5.18TVEE242 pKa = 3.96MEE244 pKa = 4.26GTNPAA249 pKa = 3.76
MM1 pKa = 7.08TGRR4 pKa = 11.84RR5 pKa = 11.84ANSSSEE11 pKa = 3.76KK12 pKa = 10.28LKK14 pKa = 11.13RR15 pKa = 11.84EE16 pKa = 3.48IANLQRR22 pKa = 11.84RR23 pKa = 11.84LATSTISSNGPRR35 pKa = 11.84SGRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84GGNGNGGGGPSVPAAMGSNPRR63 pKa = 11.84PSRR66 pKa = 11.84GRR68 pKa = 11.84RR69 pKa = 11.84GGARR73 pKa = 11.84TRR75 pKa = 11.84VGNGGRR81 pKa = 11.84IVLTRR86 pKa = 11.84DD87 pKa = 3.06EE88 pKa = 4.88LLVTVATTPDD98 pKa = 3.32KK99 pKa = 11.42NEE101 pKa = 4.27SVFSKK106 pKa = 10.91DD107 pKa = 3.61LVPSPGVMPFLFRR120 pKa = 11.84LSSCYY125 pKa = 8.83QRR127 pKa = 11.84IRR129 pKa = 11.84WVRR132 pKa = 11.84ASISWRR138 pKa = 11.84PSCGTATDD146 pKa = 5.14GIISYY151 pKa = 10.33GVAFNGSEE159 pKa = 4.32SIKK162 pKa = 10.97SRR164 pKa = 11.84DD165 pKa = 3.56LVTSLTPCNDD175 pKa = 3.01HH176 pKa = 7.08PVWQSTGISPLVIPGDD192 pKa = 3.75MLMSRR197 pKa = 11.84RR198 pKa = 11.84WYY200 pKa = 8.95PLNVTGGDD208 pKa = 3.5QFDD211 pKa = 3.46KK212 pKa = 11.4GMGKK216 pKa = 9.69FCVGLTHH223 pKa = 7.65DD224 pKa = 4.25AEE226 pKa = 4.62KK227 pKa = 10.42VAKK230 pKa = 10.22SRR232 pKa = 11.84GEE234 pKa = 3.61FWISYY239 pKa = 5.18TVEE242 pKa = 3.96MEE244 pKa = 4.26GTNPAA249 pKa = 3.76
Molecular weight: 26.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1620 |
245 |
1126 |
540.0 |
60.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.79 ± 0.737 | 0.679 ± 0.277 |
5.37 ± 0.396 | 6.79 ± 1.101 |
4.198 ± 0.56 | 7.346 ± 1.502 |
1.852 ± 0.324 | 3.272 ± 0.338 |
4.753 ± 0.617 | 8.704 ± 0.841 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.346 ± 0.177 | 3.025 ± 0.726 |
7.037 ± 0.202 | 3.333 ± 0.563 |
8.457 ± 0.955 | 7.284 ± 1.379 |
5.37 ± 0.462 | 8.457 ± 0.488 |
2.222 ± 0.068 | 2.716 ± 0.344 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
