
Fragaria chiloensis cryptic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
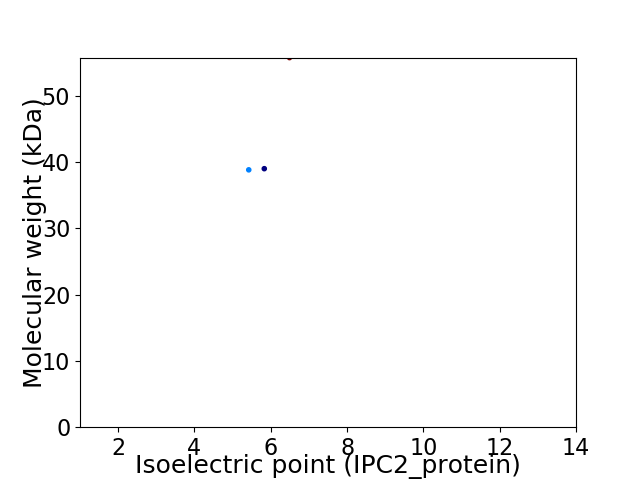
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q4FCM7|Q4FCM7_9VIRU RNA-dependent RNA polymerase OS=Fragaria chiloensis cryptic virus OX=335204 PE=4 SV=2
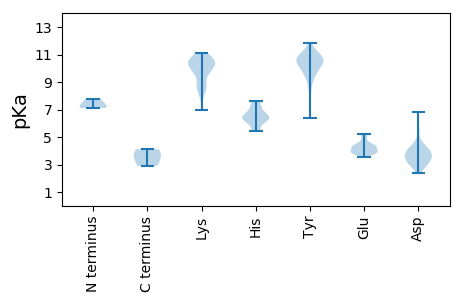
MM1 pKa = 7.28ATDD4 pKa = 3.43NSDD7 pKa = 3.71AKK9 pKa = 10.16TEE11 pKa = 3.99QSKK14 pKa = 8.04VTEE17 pKa = 4.65SKK19 pKa = 10.74GSSPTKK25 pKa = 10.25SLPAHH30 pKa = 6.32AADD33 pKa = 3.82EE34 pKa = 4.31KK35 pKa = 10.87AAVGATFKK43 pKa = 10.84LQSRR47 pKa = 11.84QPTEE51 pKa = 3.87PPAIPSVFAQSQSLNDD67 pKa = 3.67SVGNPDD73 pKa = 5.6FIAQLRR79 pKa = 11.84DD80 pKa = 2.83EE81 pKa = 4.49GMYY84 pKa = 11.23GLFLPRR90 pKa = 11.84DD91 pKa = 3.58RR92 pKa = 11.84ARR94 pKa = 11.84RR95 pKa = 11.84TPTPVTLRR103 pKa = 11.84PLTYY107 pKa = 9.25FQSIEE112 pKa = 4.06RR113 pKa = 11.84TISQSMFHH121 pKa = 7.13LLQTKK126 pKa = 8.22TEE128 pKa = 4.19FNNDD132 pKa = 3.2DD133 pKa = 4.3AIQRR137 pKa = 11.84INTIAEE143 pKa = 3.78QLAIGACLATHH154 pKa = 6.29MKK156 pKa = 10.0LRR158 pKa = 11.84ALIHH162 pKa = 6.8LDD164 pKa = 3.36FPDD167 pKa = 4.2RR168 pKa = 11.84YY169 pKa = 10.86NIGSLKK175 pKa = 10.11RR176 pKa = 11.84PKK178 pKa = 10.08TITDD182 pKa = 3.93LPVVAPFAFAIQQLGYY198 pKa = 11.3VNIANLTEE206 pKa = 3.69EE207 pKa = 3.98RR208 pKa = 11.84RR209 pKa = 11.84YY210 pKa = 10.9VPVLPEE216 pKa = 3.91TGHH219 pKa = 5.96TFGIPTGHH227 pKa = 6.23NWNPNLYY234 pKa = 9.54AQAVDD239 pKa = 3.58YY240 pKa = 10.92ARR242 pKa = 11.84KK243 pKa = 9.54FGLHH247 pKa = 6.16FNVVDD252 pKa = 3.72YY253 pKa = 9.62TKK255 pKa = 10.85KK256 pKa = 10.47QGTAWWLLRR265 pKa = 11.84QHH267 pKa = 6.62FEE269 pKa = 4.24DD270 pKa = 6.8GIFEE274 pKa = 4.4LQLPLPEE281 pKa = 4.49VNFTSSMALTLSLFLNAEE299 pKa = 4.3EE300 pKa = 4.44VNATSEE306 pKa = 4.24IFDD309 pKa = 4.23LTPVGADD316 pKa = 2.36IYY318 pKa = 10.33GYY320 pKa = 10.43IMRR323 pKa = 11.84EE324 pKa = 3.52PHH326 pKa = 6.48LGINVSTFEE335 pKa = 5.17VIDD338 pKa = 4.13EE339 pKa = 4.24SAKK342 pKa = 10.58EE343 pKa = 3.87IVSNVV348 pKa = 2.9
MM1 pKa = 7.28ATDD4 pKa = 3.43NSDD7 pKa = 3.71AKK9 pKa = 10.16TEE11 pKa = 3.99QSKK14 pKa = 8.04VTEE17 pKa = 4.65SKK19 pKa = 10.74GSSPTKK25 pKa = 10.25SLPAHH30 pKa = 6.32AADD33 pKa = 3.82EE34 pKa = 4.31KK35 pKa = 10.87AAVGATFKK43 pKa = 10.84LQSRR47 pKa = 11.84QPTEE51 pKa = 3.87PPAIPSVFAQSQSLNDD67 pKa = 3.67SVGNPDD73 pKa = 5.6FIAQLRR79 pKa = 11.84DD80 pKa = 2.83EE81 pKa = 4.49GMYY84 pKa = 11.23GLFLPRR90 pKa = 11.84DD91 pKa = 3.58RR92 pKa = 11.84ARR94 pKa = 11.84RR95 pKa = 11.84TPTPVTLRR103 pKa = 11.84PLTYY107 pKa = 9.25FQSIEE112 pKa = 4.06RR113 pKa = 11.84TISQSMFHH121 pKa = 7.13LLQTKK126 pKa = 8.22TEE128 pKa = 4.19FNNDD132 pKa = 3.2DD133 pKa = 4.3AIQRR137 pKa = 11.84INTIAEE143 pKa = 3.78QLAIGACLATHH154 pKa = 6.29MKK156 pKa = 10.0LRR158 pKa = 11.84ALIHH162 pKa = 6.8LDD164 pKa = 3.36FPDD167 pKa = 4.2RR168 pKa = 11.84YY169 pKa = 10.86NIGSLKK175 pKa = 10.11RR176 pKa = 11.84PKK178 pKa = 10.08TITDD182 pKa = 3.93LPVVAPFAFAIQQLGYY198 pKa = 11.3VNIANLTEE206 pKa = 3.69EE207 pKa = 3.98RR208 pKa = 11.84RR209 pKa = 11.84YY210 pKa = 10.9VPVLPEE216 pKa = 3.91TGHH219 pKa = 5.96TFGIPTGHH227 pKa = 6.23NWNPNLYY234 pKa = 9.54AQAVDD239 pKa = 3.58YY240 pKa = 10.92ARR242 pKa = 11.84KK243 pKa = 9.54FGLHH247 pKa = 6.16FNVVDD252 pKa = 3.72YY253 pKa = 9.62TKK255 pKa = 10.85KK256 pKa = 10.47QGTAWWLLRR265 pKa = 11.84QHH267 pKa = 6.62FEE269 pKa = 4.24DD270 pKa = 6.8GIFEE274 pKa = 4.4LQLPLPEE281 pKa = 4.49VNFTSSMALTLSLFLNAEE299 pKa = 4.3EE300 pKa = 4.44VNATSEE306 pKa = 4.24IFDD309 pKa = 4.23LTPVGADD316 pKa = 2.36IYY318 pKa = 10.33GYY320 pKa = 10.43IMRR323 pKa = 11.84EE324 pKa = 3.52PHH326 pKa = 6.48LGINVSTFEE335 pKa = 5.17VIDD338 pKa = 4.13EE339 pKa = 4.24SAKK342 pKa = 10.58EE343 pKa = 3.87IVSNVV348 pKa = 2.9
Molecular weight: 38.84 kDa
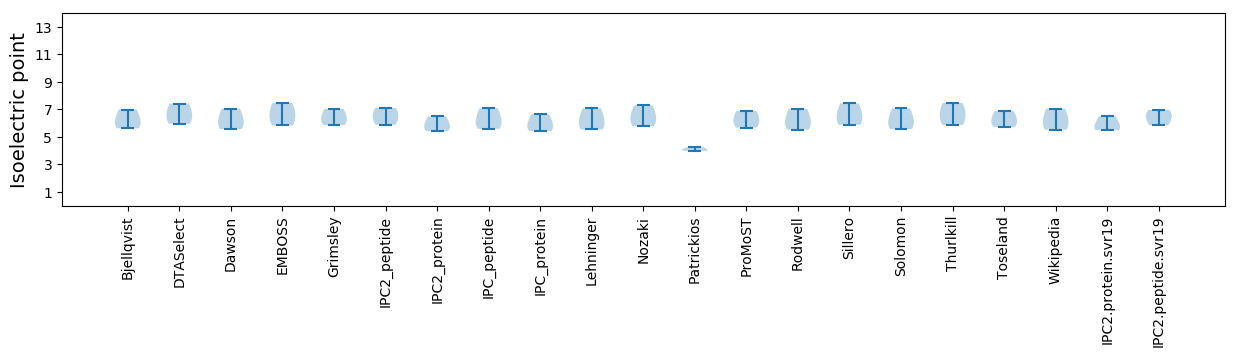
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q4FCM7|Q4FCM7_9VIRU RNA-dependent RNA polymerase OS=Fragaria chiloensis cryptic virus OX=335204 PE=4 SV=2
MM1 pKa = 7.11EE2 pKa = 4.95HH3 pKa = 7.37RR4 pKa = 11.84FRR6 pKa = 11.84GIPRR10 pKa = 11.84GLIEE14 pKa = 5.14LEE16 pKa = 4.32EE17 pKa = 4.61IPTRR21 pKa = 11.84RR22 pKa = 11.84LRR24 pKa = 11.84EE25 pKa = 3.86EE26 pKa = 3.96RR27 pKa = 11.84VVHH30 pKa = 6.15IDD32 pKa = 2.81AWSSKK37 pKa = 10.52AIDD40 pKa = 4.66AIVPLSLRR48 pKa = 11.84IEE50 pKa = 4.27LDD52 pKa = 2.72GWARR56 pKa = 11.84SYY58 pKa = 10.12YY59 pKa = 9.73TLQAHH64 pKa = 7.29IDD66 pKa = 3.69SIMQYY71 pKa = 10.73DD72 pKa = 4.34RR73 pKa = 11.84PKK75 pKa = 10.83LPQPTNAAWNTTTQHH90 pKa = 5.49VRR92 pKa = 11.84TQFARR97 pKa = 11.84MDD99 pKa = 3.5KK100 pKa = 10.52VQTLSYY106 pKa = 10.28LQLDD110 pKa = 3.44QVKK113 pKa = 8.55WVRR116 pKa = 11.84SSAAGYY122 pKa = 10.1GYY124 pKa = 10.84VGRR127 pKa = 11.84KK128 pKa = 8.82SDD130 pKa = 3.29NDD132 pKa = 3.18NYY134 pKa = 10.4FRR136 pKa = 11.84ARR138 pKa = 11.84KK139 pKa = 6.97TAFTIAEE146 pKa = 4.41KK147 pKa = 10.75LNHH150 pKa = 6.4DD151 pKa = 3.96RR152 pKa = 11.84DD153 pKa = 4.13YY154 pKa = 11.81GPLALEE160 pKa = 5.24DD161 pKa = 4.09STPDD165 pKa = 2.95IAFTRR170 pKa = 11.84TQLCQIKK177 pKa = 10.09VKK179 pKa = 10.68RR180 pKa = 11.84KK181 pKa = 8.85IRR183 pKa = 11.84NVWGEE188 pKa = 3.7AFHH191 pKa = 6.65YY192 pKa = 10.62VLLEE196 pKa = 3.85GLFADD201 pKa = 5.87PIIQHH206 pKa = 6.73FIRR209 pKa = 11.84NKK211 pKa = 8.32SFYY214 pKa = 10.66FIGEE218 pKa = 4.28DD219 pKa = 3.35PLLAVPRR226 pKa = 11.84LVEE229 pKa = 4.85KK230 pKa = 10.51ILSEE234 pKa = 3.71QDD236 pKa = 3.3YY237 pKa = 11.07VYY239 pKa = 10.61MFDD242 pKa = 3.19WSGFDD247 pKa = 3.73ASVQEE252 pKa = 4.0WEE254 pKa = 3.77IRR256 pKa = 11.84FAFSLLEE263 pKa = 4.4SILIFPSSVEE273 pKa = 3.88SYY275 pKa = 8.34IWHH278 pKa = 6.89FIIEE282 pKa = 4.06LFIYY286 pKa = 10.16RR287 pKa = 11.84KK288 pKa = 9.36IAAPNGKK295 pKa = 9.81VYY297 pKa = 10.9LKK299 pKa = 9.95TLGIPSGSCFTNIIGSIVNYY319 pKa = 10.14VRR321 pKa = 11.84IQYY324 pKa = 10.66LFFRR328 pKa = 11.84LTNNFVTVFTHH339 pKa = 7.41GDD341 pKa = 3.4DD342 pKa = 3.78SLVGVSTTQYY352 pKa = 9.38VQMDD356 pKa = 4.15NFEE359 pKa = 5.14PICAEE364 pKa = 4.14HH365 pKa = 6.52NWTINIAKK373 pKa = 9.99SAVSHH378 pKa = 5.51EE379 pKa = 4.3AEE381 pKa = 3.98GVSFLSRR388 pKa = 11.84KK389 pKa = 8.61VRR391 pKa = 11.84EE392 pKa = 4.09HH393 pKa = 5.41CHH395 pKa = 6.31ARR397 pKa = 11.84DD398 pKa = 3.3EE399 pKa = 4.34LLCLRR404 pKa = 11.84MLKK407 pKa = 10.32FPEE410 pKa = 4.13YY411 pKa = 9.64VVEE414 pKa = 4.33SGAMSTLRR422 pKa = 11.84AHH424 pKa = 7.62SIHH427 pKa = 6.47QDD429 pKa = 2.59AGINSRR435 pKa = 11.84YY436 pKa = 9.78LYY438 pKa = 10.51SIYY441 pKa = 10.45KK442 pKa = 9.67YY443 pKa = 10.62LLHH446 pKa = 7.45RR447 pKa = 11.84YY448 pKa = 8.49GKK450 pKa = 10.52ADD452 pKa = 3.81SLPLNQQNWDD462 pKa = 3.52PLEE465 pKa = 4.09YY466 pKa = 10.65EE467 pKa = 3.99NLRR470 pKa = 11.84VSFATQNYY478 pKa = 6.39EE479 pKa = 3.61
MM1 pKa = 7.11EE2 pKa = 4.95HH3 pKa = 7.37RR4 pKa = 11.84FRR6 pKa = 11.84GIPRR10 pKa = 11.84GLIEE14 pKa = 5.14LEE16 pKa = 4.32EE17 pKa = 4.61IPTRR21 pKa = 11.84RR22 pKa = 11.84LRR24 pKa = 11.84EE25 pKa = 3.86EE26 pKa = 3.96RR27 pKa = 11.84VVHH30 pKa = 6.15IDD32 pKa = 2.81AWSSKK37 pKa = 10.52AIDD40 pKa = 4.66AIVPLSLRR48 pKa = 11.84IEE50 pKa = 4.27LDD52 pKa = 2.72GWARR56 pKa = 11.84SYY58 pKa = 10.12YY59 pKa = 9.73TLQAHH64 pKa = 7.29IDD66 pKa = 3.69SIMQYY71 pKa = 10.73DD72 pKa = 4.34RR73 pKa = 11.84PKK75 pKa = 10.83LPQPTNAAWNTTTQHH90 pKa = 5.49VRR92 pKa = 11.84TQFARR97 pKa = 11.84MDD99 pKa = 3.5KK100 pKa = 10.52VQTLSYY106 pKa = 10.28LQLDD110 pKa = 3.44QVKK113 pKa = 8.55WVRR116 pKa = 11.84SSAAGYY122 pKa = 10.1GYY124 pKa = 10.84VGRR127 pKa = 11.84KK128 pKa = 8.82SDD130 pKa = 3.29NDD132 pKa = 3.18NYY134 pKa = 10.4FRR136 pKa = 11.84ARR138 pKa = 11.84KK139 pKa = 6.97TAFTIAEE146 pKa = 4.41KK147 pKa = 10.75LNHH150 pKa = 6.4DD151 pKa = 3.96RR152 pKa = 11.84DD153 pKa = 4.13YY154 pKa = 11.81GPLALEE160 pKa = 5.24DD161 pKa = 4.09STPDD165 pKa = 2.95IAFTRR170 pKa = 11.84TQLCQIKK177 pKa = 10.09VKK179 pKa = 10.68RR180 pKa = 11.84KK181 pKa = 8.85IRR183 pKa = 11.84NVWGEE188 pKa = 3.7AFHH191 pKa = 6.65YY192 pKa = 10.62VLLEE196 pKa = 3.85GLFADD201 pKa = 5.87PIIQHH206 pKa = 6.73FIRR209 pKa = 11.84NKK211 pKa = 8.32SFYY214 pKa = 10.66FIGEE218 pKa = 4.28DD219 pKa = 3.35PLLAVPRR226 pKa = 11.84LVEE229 pKa = 4.85KK230 pKa = 10.51ILSEE234 pKa = 3.71QDD236 pKa = 3.3YY237 pKa = 11.07VYY239 pKa = 10.61MFDD242 pKa = 3.19WSGFDD247 pKa = 3.73ASVQEE252 pKa = 4.0WEE254 pKa = 3.77IRR256 pKa = 11.84FAFSLLEE263 pKa = 4.4SILIFPSSVEE273 pKa = 3.88SYY275 pKa = 8.34IWHH278 pKa = 6.89FIIEE282 pKa = 4.06LFIYY286 pKa = 10.16RR287 pKa = 11.84KK288 pKa = 9.36IAAPNGKK295 pKa = 9.81VYY297 pKa = 10.9LKK299 pKa = 9.95TLGIPSGSCFTNIIGSIVNYY319 pKa = 10.14VRR321 pKa = 11.84IQYY324 pKa = 10.66LFFRR328 pKa = 11.84LTNNFVTVFTHH339 pKa = 7.41GDD341 pKa = 3.4DD342 pKa = 3.78SLVGVSTTQYY352 pKa = 9.38VQMDD356 pKa = 4.15NFEE359 pKa = 5.14PICAEE364 pKa = 4.14HH365 pKa = 6.52NWTINIAKK373 pKa = 9.99SAVSHH378 pKa = 5.51EE379 pKa = 4.3AEE381 pKa = 3.98GVSFLSRR388 pKa = 11.84KK389 pKa = 8.61VRR391 pKa = 11.84EE392 pKa = 4.09HH393 pKa = 5.41CHH395 pKa = 6.31ARR397 pKa = 11.84DD398 pKa = 3.3EE399 pKa = 4.34LLCLRR404 pKa = 11.84MLKK407 pKa = 10.32FPEE410 pKa = 4.13YY411 pKa = 9.64VVEE414 pKa = 4.33SGAMSTLRR422 pKa = 11.84AHH424 pKa = 7.62SIHH427 pKa = 6.47QDD429 pKa = 2.59AGINSRR435 pKa = 11.84YY436 pKa = 9.78LYY438 pKa = 10.51SIYY441 pKa = 10.45KK442 pKa = 9.67YY443 pKa = 10.62LLHH446 pKa = 7.45RR447 pKa = 11.84YY448 pKa = 8.49GKK450 pKa = 10.52ADD452 pKa = 3.81SLPLNQQNWDD462 pKa = 3.52PLEE465 pKa = 4.09YY466 pKa = 10.65EE467 pKa = 3.99NLRR470 pKa = 11.84VSFATQNYY478 pKa = 6.39EE479 pKa = 3.61
Molecular weight: 55.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1173 |
346 |
479 |
391.0 |
44.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.355 ± 0.867 | 1.108 ± 0.395 |
5.286 ± 0.08 | 6.223 ± 0.075 |
4.604 ± 0.773 | 4.263 ± 0.328 |
2.899 ± 0.214 | 7.076 ± 0.406 |
4.007 ± 0.103 | 9.037 ± 0.262 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.876 ± 0.288 | 4.689 ± 0.254 |
5.541 ± 0.872 | 4.007 ± 0.461 |
6.394 ± 0.564 | 6.479 ± 0.45 |
6.564 ± 0.859 | 5.797 ± 0.262 |
1.449 ± 0.314 | 4.348 ± 0.608 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
