
Rodent Torque teno virus 7
Taxonomy: Viruses; Anelloviridae; Wawtorquevirus; Torque teno rodent virus 3
Average proteome isoelectric point is 8.81
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4QBC6|A0A2H4QBC6_9VIRU Capsid protein OS=Rodent Torque teno virus 7 OX=2054614 PE=3 SV=1
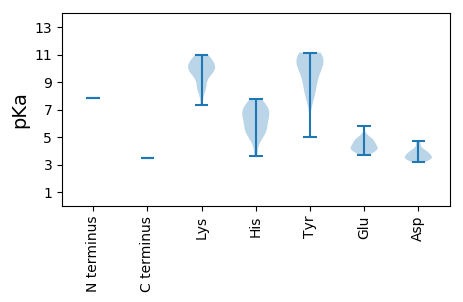
MM1 pKa = 7.87AYY3 pKa = 10.42YY4 pKa = 10.32SYY6 pKa = 10.62RR7 pKa = 11.84RR8 pKa = 11.84NWRR11 pKa = 11.84RR12 pKa = 11.84PFPRR16 pKa = 11.84RR17 pKa = 11.84GGYY20 pKa = 5.01WTRR23 pKa = 11.84RR24 pKa = 11.84LWTRR28 pKa = 11.84AYY30 pKa = 8.93HH31 pKa = 5.71RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84GYY37 pKa = 8.19YY38 pKa = 7.42RR39 pKa = 11.84TYY41 pKa = 9.39RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84WRR46 pKa = 11.84PVRR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 10.06RR52 pKa = 11.84NYY54 pKa = 9.13HH55 pKa = 5.58PRR57 pKa = 11.84KK58 pKa = 8.54HH59 pKa = 5.55WVPVPQWNPSNKK71 pKa = 8.58RR72 pKa = 11.84WCTIRR77 pKa = 11.84GYY79 pKa = 9.77MPLLFCVGAPAGCAVFNYY97 pKa = 8.15PQKK100 pKa = 9.76TVSDD104 pKa = 3.59WKK106 pKa = 10.96GGGVHH111 pKa = 6.65SSVFSLADD119 pKa = 4.07LYY121 pKa = 11.14QEE123 pKa = 4.05EE124 pKa = 4.96TEE126 pKa = 4.34WRR128 pKa = 11.84CRR130 pKa = 11.84WSGSNQGFNLCRR142 pKa = 11.84YY143 pKa = 9.71YY144 pKa = 11.13GGTLCLWPNATYY156 pKa = 10.47SYY158 pKa = 10.64IFWYY162 pKa = 10.14SVNDD166 pKa = 4.1PEE168 pKa = 5.79PDD170 pKa = 3.45NLPLRR175 pKa = 11.84VCHH178 pKa = 6.35PSQMLVQPRR187 pKa = 11.84HH188 pKa = 5.13IVVTRR193 pKa = 11.84ASMHH197 pKa = 6.75RR198 pKa = 11.84NMKK201 pKa = 8.75PIKK204 pKa = 10.47VRR206 pKa = 11.84IPPPATLAGNWYY218 pKa = 10.35SMKK221 pKa = 10.91DD222 pKa = 3.38MCQKK226 pKa = 9.34TLFKK230 pKa = 10.39WRR232 pKa = 11.84VSLIDD237 pKa = 4.7MEE239 pKa = 4.54QTWTGFPDD247 pKa = 3.28GHH249 pKa = 7.75APVSAQVFACRR260 pKa = 11.84PGQEE264 pKa = 4.57ALTQTVLSYY273 pKa = 10.69YY274 pKa = 10.01PWLDD278 pKa = 3.49DD279 pKa = 3.92GNNVNISEE287 pKa = 4.25HH288 pKa = 4.73TMEE291 pKa = 4.78FNQTNSRR298 pKa = 11.84PTKK301 pKa = 7.3TANYY305 pKa = 7.9NWPIEE310 pKa = 3.98AQFRR314 pKa = 11.84KK315 pKa = 10.11LLLPFYY321 pKa = 8.91MWGFGFPTEE330 pKa = 4.27YY331 pKa = 11.11YY332 pKa = 8.66IDD334 pKa = 3.79SYY336 pKa = 11.03QWHH339 pKa = 7.08LPAPHH344 pKa = 7.17LDD346 pKa = 3.43SHH348 pKa = 6.59NKK350 pKa = 9.71NVMSFIFLSIKK361 pKa = 10.53AGAHH365 pKa = 3.61QWKK368 pKa = 9.88GFDD371 pKa = 3.51TTNVYY376 pKa = 9.83YY377 pKa = 11.13VPFSTLLKK385 pKa = 9.86IVCNAPWVEE394 pKa = 5.04KK395 pKa = 10.52SIQQPGVNVTMMYY408 pKa = 10.68KK409 pKa = 10.48FYY411 pKa = 10.12FQWGGKK417 pKa = 8.52PGSKK421 pKa = 10.03LPPVTPCDD429 pKa = 3.85PSAADD434 pKa = 3.31YY435 pKa = 11.02KK436 pKa = 10.89PIIEE440 pKa = 4.8HH441 pKa = 7.2PYY443 pKa = 9.16TYY445 pKa = 10.29DD446 pKa = 3.62AEE448 pKa = 4.63VPGPGDD454 pKa = 3.57YY455 pKa = 11.05DD456 pKa = 3.82SDD458 pKa = 4.49GIIRR462 pKa = 11.84EE463 pKa = 4.09GALRR467 pKa = 11.84RR468 pKa = 11.84LTRR471 pKa = 11.84SNIPTDD477 pKa = 3.16ARR479 pKa = 11.84KK480 pKa = 9.79LSRR483 pKa = 11.84SALHH487 pKa = 5.66QSAIAAYY494 pKa = 8.21RR495 pKa = 11.84TEE497 pKa = 4.75HH498 pKa = 6.4GPSSGEE504 pKa = 4.04SEE506 pKa = 4.29QEE508 pKa = 4.19EE509 pKa = 4.73EE510 pKa = 4.01DD511 pKa = 3.95TPYY514 pKa = 10.98LSCSSEE520 pKa = 4.06EE521 pKa = 4.23SQKK524 pKa = 9.86DD525 pKa = 3.69TPSSSVRR532 pKa = 11.84QRR534 pKa = 11.84LRR536 pKa = 11.84EE537 pKa = 3.66RR538 pKa = 11.84LGRR541 pKa = 11.84MDD543 pKa = 3.39RR544 pKa = 11.84HH545 pKa = 5.51RR546 pKa = 11.84LHH548 pKa = 7.45RR549 pKa = 11.84ILRR552 pKa = 11.84LLKK555 pKa = 9.84SGHH558 pKa = 6.8RR559 pKa = 11.84IHH561 pKa = 7.08TSTHH565 pKa = 5.13HH566 pKa = 6.17SGRR569 pKa = 11.84EE570 pKa = 3.67RR571 pKa = 11.84SSSII575 pKa = 3.47
MM1 pKa = 7.87AYY3 pKa = 10.42YY4 pKa = 10.32SYY6 pKa = 10.62RR7 pKa = 11.84RR8 pKa = 11.84NWRR11 pKa = 11.84RR12 pKa = 11.84PFPRR16 pKa = 11.84RR17 pKa = 11.84GGYY20 pKa = 5.01WTRR23 pKa = 11.84RR24 pKa = 11.84LWTRR28 pKa = 11.84AYY30 pKa = 8.93HH31 pKa = 5.71RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84GYY37 pKa = 8.19YY38 pKa = 7.42RR39 pKa = 11.84TYY41 pKa = 9.39RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84WRR46 pKa = 11.84PVRR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 10.06RR52 pKa = 11.84NYY54 pKa = 9.13HH55 pKa = 5.58PRR57 pKa = 11.84KK58 pKa = 8.54HH59 pKa = 5.55WVPVPQWNPSNKK71 pKa = 8.58RR72 pKa = 11.84WCTIRR77 pKa = 11.84GYY79 pKa = 9.77MPLLFCVGAPAGCAVFNYY97 pKa = 8.15PQKK100 pKa = 9.76TVSDD104 pKa = 3.59WKK106 pKa = 10.96GGGVHH111 pKa = 6.65SSVFSLADD119 pKa = 4.07LYY121 pKa = 11.14QEE123 pKa = 4.05EE124 pKa = 4.96TEE126 pKa = 4.34WRR128 pKa = 11.84CRR130 pKa = 11.84WSGSNQGFNLCRR142 pKa = 11.84YY143 pKa = 9.71YY144 pKa = 11.13GGTLCLWPNATYY156 pKa = 10.47SYY158 pKa = 10.64IFWYY162 pKa = 10.14SVNDD166 pKa = 4.1PEE168 pKa = 5.79PDD170 pKa = 3.45NLPLRR175 pKa = 11.84VCHH178 pKa = 6.35PSQMLVQPRR187 pKa = 11.84HH188 pKa = 5.13IVVTRR193 pKa = 11.84ASMHH197 pKa = 6.75RR198 pKa = 11.84NMKK201 pKa = 8.75PIKK204 pKa = 10.47VRR206 pKa = 11.84IPPPATLAGNWYY218 pKa = 10.35SMKK221 pKa = 10.91DD222 pKa = 3.38MCQKK226 pKa = 9.34TLFKK230 pKa = 10.39WRR232 pKa = 11.84VSLIDD237 pKa = 4.7MEE239 pKa = 4.54QTWTGFPDD247 pKa = 3.28GHH249 pKa = 7.75APVSAQVFACRR260 pKa = 11.84PGQEE264 pKa = 4.57ALTQTVLSYY273 pKa = 10.69YY274 pKa = 10.01PWLDD278 pKa = 3.49DD279 pKa = 3.92GNNVNISEE287 pKa = 4.25HH288 pKa = 4.73TMEE291 pKa = 4.78FNQTNSRR298 pKa = 11.84PTKK301 pKa = 7.3TANYY305 pKa = 7.9NWPIEE310 pKa = 3.98AQFRR314 pKa = 11.84KK315 pKa = 10.11LLLPFYY321 pKa = 8.91MWGFGFPTEE330 pKa = 4.27YY331 pKa = 11.11YY332 pKa = 8.66IDD334 pKa = 3.79SYY336 pKa = 11.03QWHH339 pKa = 7.08LPAPHH344 pKa = 7.17LDD346 pKa = 3.43SHH348 pKa = 6.59NKK350 pKa = 9.71NVMSFIFLSIKK361 pKa = 10.53AGAHH365 pKa = 3.61QWKK368 pKa = 9.88GFDD371 pKa = 3.51TTNVYY376 pKa = 9.83YY377 pKa = 11.13VPFSTLLKK385 pKa = 9.86IVCNAPWVEE394 pKa = 5.04KK395 pKa = 10.52SIQQPGVNVTMMYY408 pKa = 10.68KK409 pKa = 10.48FYY411 pKa = 10.12FQWGGKK417 pKa = 8.52PGSKK421 pKa = 10.03LPPVTPCDD429 pKa = 3.85PSAADD434 pKa = 3.31YY435 pKa = 11.02KK436 pKa = 10.89PIIEE440 pKa = 4.8HH441 pKa = 7.2PYY443 pKa = 9.16TYY445 pKa = 10.29DD446 pKa = 3.62AEE448 pKa = 4.63VPGPGDD454 pKa = 3.57YY455 pKa = 11.05DD456 pKa = 3.82SDD458 pKa = 4.49GIIRR462 pKa = 11.84EE463 pKa = 4.09GALRR467 pKa = 11.84RR468 pKa = 11.84LTRR471 pKa = 11.84SNIPTDD477 pKa = 3.16ARR479 pKa = 11.84KK480 pKa = 9.79LSRR483 pKa = 11.84SALHH487 pKa = 5.66QSAIAAYY494 pKa = 8.21RR495 pKa = 11.84TEE497 pKa = 4.75HH498 pKa = 6.4GPSSGEE504 pKa = 4.04SEE506 pKa = 4.29QEE508 pKa = 4.19EE509 pKa = 4.73EE510 pKa = 4.01DD511 pKa = 3.95TPYY514 pKa = 10.98LSCSSEE520 pKa = 4.06EE521 pKa = 4.23SQKK524 pKa = 9.86DD525 pKa = 3.69TPSSSVRR532 pKa = 11.84QRR534 pKa = 11.84LRR536 pKa = 11.84EE537 pKa = 3.66RR538 pKa = 11.84LGRR541 pKa = 11.84MDD543 pKa = 3.39RR544 pKa = 11.84HH545 pKa = 5.51RR546 pKa = 11.84LHH548 pKa = 7.45RR549 pKa = 11.84ILRR552 pKa = 11.84LLKK555 pKa = 9.84SGHH558 pKa = 6.8RR559 pKa = 11.84IHH561 pKa = 7.08TSTHH565 pKa = 5.13HH566 pKa = 6.17SGRR569 pKa = 11.84EE570 pKa = 3.67RR571 pKa = 11.84SSSII575 pKa = 3.47
Molecular weight: 67.34 kDa
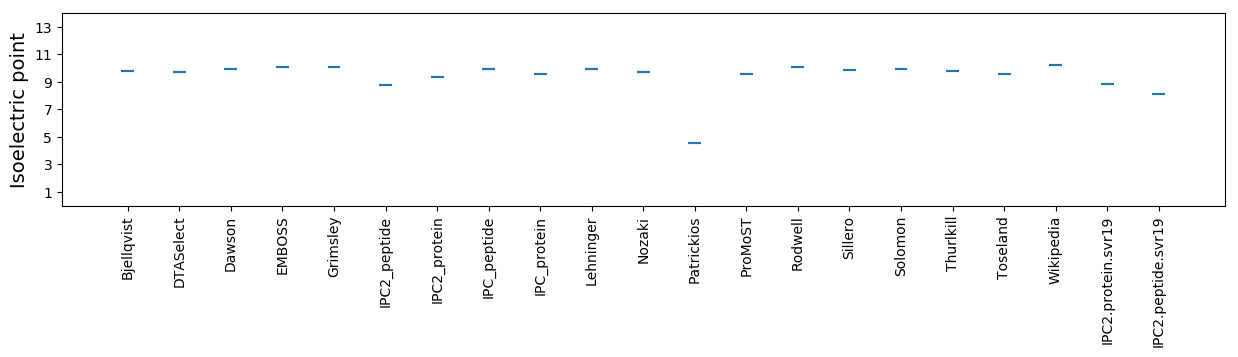
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4QBC6|A0A2H4QBC6_9VIRU Capsid protein OS=Rodent Torque teno virus 7 OX=2054614 PE=3 SV=1
MM1 pKa = 7.87AYY3 pKa = 10.42YY4 pKa = 10.32SYY6 pKa = 10.62RR7 pKa = 11.84RR8 pKa = 11.84NWRR11 pKa = 11.84RR12 pKa = 11.84PFPRR16 pKa = 11.84RR17 pKa = 11.84GGYY20 pKa = 5.01WTRR23 pKa = 11.84RR24 pKa = 11.84LWTRR28 pKa = 11.84AYY30 pKa = 8.93HH31 pKa = 5.71RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84GYY37 pKa = 8.19YY38 pKa = 7.42RR39 pKa = 11.84TYY41 pKa = 9.39RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84WRR46 pKa = 11.84PVRR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 10.06RR52 pKa = 11.84NYY54 pKa = 9.13HH55 pKa = 5.58PRR57 pKa = 11.84KK58 pKa = 8.54HH59 pKa = 5.55WVPVPQWNPSNKK71 pKa = 8.58RR72 pKa = 11.84WCTIRR77 pKa = 11.84GYY79 pKa = 9.77MPLLFCVGAPAGCAVFNYY97 pKa = 8.15PQKK100 pKa = 9.76TVSDD104 pKa = 3.59WKK106 pKa = 10.96GGGVHH111 pKa = 6.65SSVFSLADD119 pKa = 4.07LYY121 pKa = 11.14QEE123 pKa = 4.05EE124 pKa = 4.96TEE126 pKa = 4.34WRR128 pKa = 11.84CRR130 pKa = 11.84WSGSNQGFNLCRR142 pKa = 11.84YY143 pKa = 9.71YY144 pKa = 11.13GGTLCLWPNATYY156 pKa = 10.47SYY158 pKa = 10.64IFWYY162 pKa = 10.14SVNDD166 pKa = 4.1PEE168 pKa = 5.79PDD170 pKa = 3.45NLPLRR175 pKa = 11.84VCHH178 pKa = 6.35PSQMLVQPRR187 pKa = 11.84HH188 pKa = 5.13IVVTRR193 pKa = 11.84ASMHH197 pKa = 6.75RR198 pKa = 11.84NMKK201 pKa = 8.75PIKK204 pKa = 10.47VRR206 pKa = 11.84IPPPATLAGNWYY218 pKa = 10.35SMKK221 pKa = 10.91DD222 pKa = 3.38MCQKK226 pKa = 9.34TLFKK230 pKa = 10.39WRR232 pKa = 11.84VSLIDD237 pKa = 4.7MEE239 pKa = 4.54QTWTGFPDD247 pKa = 3.28GHH249 pKa = 7.75APVSAQVFACRR260 pKa = 11.84PGQEE264 pKa = 4.57ALTQTVLSYY273 pKa = 10.69YY274 pKa = 10.01PWLDD278 pKa = 3.49DD279 pKa = 3.92GNNVNISEE287 pKa = 4.25HH288 pKa = 4.73TMEE291 pKa = 4.78FNQTNSRR298 pKa = 11.84PTKK301 pKa = 7.3TANYY305 pKa = 7.9NWPIEE310 pKa = 3.98AQFRR314 pKa = 11.84KK315 pKa = 10.11LLLPFYY321 pKa = 8.91MWGFGFPTEE330 pKa = 4.27YY331 pKa = 11.11YY332 pKa = 8.66IDD334 pKa = 3.79SYY336 pKa = 11.03QWHH339 pKa = 7.08LPAPHH344 pKa = 7.17LDD346 pKa = 3.43SHH348 pKa = 6.59NKK350 pKa = 9.71NVMSFIFLSIKK361 pKa = 10.53AGAHH365 pKa = 3.61QWKK368 pKa = 9.88GFDD371 pKa = 3.51TTNVYY376 pKa = 9.83YY377 pKa = 11.13VPFSTLLKK385 pKa = 9.86IVCNAPWVEE394 pKa = 5.04KK395 pKa = 10.52SIQQPGVNVTMMYY408 pKa = 10.68KK409 pKa = 10.48FYY411 pKa = 10.12FQWGGKK417 pKa = 8.52PGSKK421 pKa = 10.03LPPVTPCDD429 pKa = 3.85PSAADD434 pKa = 3.31YY435 pKa = 11.02KK436 pKa = 10.89PIIEE440 pKa = 4.8HH441 pKa = 7.2PYY443 pKa = 9.16TYY445 pKa = 10.29DD446 pKa = 3.62AEE448 pKa = 4.63VPGPGDD454 pKa = 3.57YY455 pKa = 11.05DD456 pKa = 3.82SDD458 pKa = 4.49GIIRR462 pKa = 11.84EE463 pKa = 4.09GALRR467 pKa = 11.84RR468 pKa = 11.84LTRR471 pKa = 11.84SNIPTDD477 pKa = 3.16ARR479 pKa = 11.84KK480 pKa = 9.79LSRR483 pKa = 11.84SALHH487 pKa = 5.66QSAIAAYY494 pKa = 8.21RR495 pKa = 11.84TEE497 pKa = 4.75HH498 pKa = 6.4GPSSGEE504 pKa = 4.04SEE506 pKa = 4.29QEE508 pKa = 4.19EE509 pKa = 4.73EE510 pKa = 4.01DD511 pKa = 3.95TPYY514 pKa = 10.98LSCSSEE520 pKa = 4.06EE521 pKa = 4.23SQKK524 pKa = 9.86DD525 pKa = 3.69TPSSSVRR532 pKa = 11.84QRR534 pKa = 11.84LRR536 pKa = 11.84EE537 pKa = 3.66RR538 pKa = 11.84LGRR541 pKa = 11.84MDD543 pKa = 3.39RR544 pKa = 11.84HH545 pKa = 5.51RR546 pKa = 11.84LHH548 pKa = 7.45RR549 pKa = 11.84ILRR552 pKa = 11.84LLKK555 pKa = 9.84SGHH558 pKa = 6.8RR559 pKa = 11.84IHH561 pKa = 7.08TSTHH565 pKa = 5.13HH566 pKa = 6.17SGRR569 pKa = 11.84EE570 pKa = 3.67RR571 pKa = 11.84SSSII575 pKa = 3.47
MM1 pKa = 7.87AYY3 pKa = 10.42YY4 pKa = 10.32SYY6 pKa = 10.62RR7 pKa = 11.84RR8 pKa = 11.84NWRR11 pKa = 11.84RR12 pKa = 11.84PFPRR16 pKa = 11.84RR17 pKa = 11.84GGYY20 pKa = 5.01WTRR23 pKa = 11.84RR24 pKa = 11.84LWTRR28 pKa = 11.84AYY30 pKa = 8.93HH31 pKa = 5.71RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84GYY37 pKa = 8.19YY38 pKa = 7.42RR39 pKa = 11.84TYY41 pKa = 9.39RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84WRR46 pKa = 11.84PVRR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 10.06RR52 pKa = 11.84NYY54 pKa = 9.13HH55 pKa = 5.58PRR57 pKa = 11.84KK58 pKa = 8.54HH59 pKa = 5.55WVPVPQWNPSNKK71 pKa = 8.58RR72 pKa = 11.84WCTIRR77 pKa = 11.84GYY79 pKa = 9.77MPLLFCVGAPAGCAVFNYY97 pKa = 8.15PQKK100 pKa = 9.76TVSDD104 pKa = 3.59WKK106 pKa = 10.96GGGVHH111 pKa = 6.65SSVFSLADD119 pKa = 4.07LYY121 pKa = 11.14QEE123 pKa = 4.05EE124 pKa = 4.96TEE126 pKa = 4.34WRR128 pKa = 11.84CRR130 pKa = 11.84WSGSNQGFNLCRR142 pKa = 11.84YY143 pKa = 9.71YY144 pKa = 11.13GGTLCLWPNATYY156 pKa = 10.47SYY158 pKa = 10.64IFWYY162 pKa = 10.14SVNDD166 pKa = 4.1PEE168 pKa = 5.79PDD170 pKa = 3.45NLPLRR175 pKa = 11.84VCHH178 pKa = 6.35PSQMLVQPRR187 pKa = 11.84HH188 pKa = 5.13IVVTRR193 pKa = 11.84ASMHH197 pKa = 6.75RR198 pKa = 11.84NMKK201 pKa = 8.75PIKK204 pKa = 10.47VRR206 pKa = 11.84IPPPATLAGNWYY218 pKa = 10.35SMKK221 pKa = 10.91DD222 pKa = 3.38MCQKK226 pKa = 9.34TLFKK230 pKa = 10.39WRR232 pKa = 11.84VSLIDD237 pKa = 4.7MEE239 pKa = 4.54QTWTGFPDD247 pKa = 3.28GHH249 pKa = 7.75APVSAQVFACRR260 pKa = 11.84PGQEE264 pKa = 4.57ALTQTVLSYY273 pKa = 10.69YY274 pKa = 10.01PWLDD278 pKa = 3.49DD279 pKa = 3.92GNNVNISEE287 pKa = 4.25HH288 pKa = 4.73TMEE291 pKa = 4.78FNQTNSRR298 pKa = 11.84PTKK301 pKa = 7.3TANYY305 pKa = 7.9NWPIEE310 pKa = 3.98AQFRR314 pKa = 11.84KK315 pKa = 10.11LLLPFYY321 pKa = 8.91MWGFGFPTEE330 pKa = 4.27YY331 pKa = 11.11YY332 pKa = 8.66IDD334 pKa = 3.79SYY336 pKa = 11.03QWHH339 pKa = 7.08LPAPHH344 pKa = 7.17LDD346 pKa = 3.43SHH348 pKa = 6.59NKK350 pKa = 9.71NVMSFIFLSIKK361 pKa = 10.53AGAHH365 pKa = 3.61QWKK368 pKa = 9.88GFDD371 pKa = 3.51TTNVYY376 pKa = 9.83YY377 pKa = 11.13VPFSTLLKK385 pKa = 9.86IVCNAPWVEE394 pKa = 5.04KK395 pKa = 10.52SIQQPGVNVTMMYY408 pKa = 10.68KK409 pKa = 10.48FYY411 pKa = 10.12FQWGGKK417 pKa = 8.52PGSKK421 pKa = 10.03LPPVTPCDD429 pKa = 3.85PSAADD434 pKa = 3.31YY435 pKa = 11.02KK436 pKa = 10.89PIIEE440 pKa = 4.8HH441 pKa = 7.2PYY443 pKa = 9.16TYY445 pKa = 10.29DD446 pKa = 3.62AEE448 pKa = 4.63VPGPGDD454 pKa = 3.57YY455 pKa = 11.05DD456 pKa = 3.82SDD458 pKa = 4.49GIIRR462 pKa = 11.84EE463 pKa = 4.09GALRR467 pKa = 11.84RR468 pKa = 11.84LTRR471 pKa = 11.84SNIPTDD477 pKa = 3.16ARR479 pKa = 11.84KK480 pKa = 9.79LSRR483 pKa = 11.84SALHH487 pKa = 5.66QSAIAAYY494 pKa = 8.21RR495 pKa = 11.84TEE497 pKa = 4.75HH498 pKa = 6.4GPSSGEE504 pKa = 4.04SEE506 pKa = 4.29QEE508 pKa = 4.19EE509 pKa = 4.73EE510 pKa = 4.01DD511 pKa = 3.95TPYY514 pKa = 10.98LSCSSEE520 pKa = 4.06EE521 pKa = 4.23SQKK524 pKa = 9.86DD525 pKa = 3.69TPSSSVRR532 pKa = 11.84QRR534 pKa = 11.84LRR536 pKa = 11.84EE537 pKa = 3.66RR538 pKa = 11.84LGRR541 pKa = 11.84MDD543 pKa = 3.39RR544 pKa = 11.84HH545 pKa = 5.51RR546 pKa = 11.84LHH548 pKa = 7.45RR549 pKa = 11.84ILRR552 pKa = 11.84LLKK555 pKa = 9.84SGHH558 pKa = 6.8RR559 pKa = 11.84IHH561 pKa = 7.08TSTHH565 pKa = 5.13HH566 pKa = 6.17SGRR569 pKa = 11.84EE570 pKa = 3.67RR571 pKa = 11.84SSSII575 pKa = 3.47
Molecular weight: 67.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
575 |
575 |
575 |
575.0 |
67.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.043 ± 0.0 | 2.087 ± 0.0 |
3.826 ± 0.0 | 4.174 ± 0.0 |
3.478 ± 0.0 | 6.087 ± 0.0 |
3.826 ± 0.0 | 3.826 ± 0.0 |
4.174 ± 0.0 | 6.435 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.435 ± 0.0 | 4.348 ± 0.0 |
8.0 ± 0.0 | 3.826 ± 0.0 |
9.565 ± 0.0 | 8.174 ± 0.0 |
5.739 ± 0.0 | 5.043 ± 0.0 |
3.826 ± 0.0 | 6.087 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
