
filamentous cyanobacterium CCP2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; unclassified Cyanobacteria
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
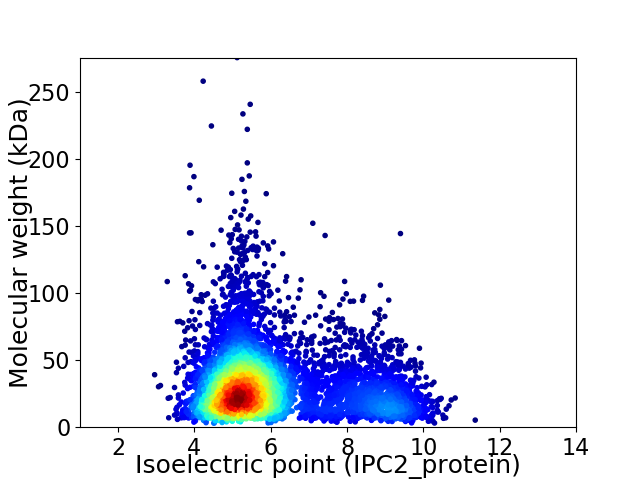
Virtual 2D-PAGE plot for 6213 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
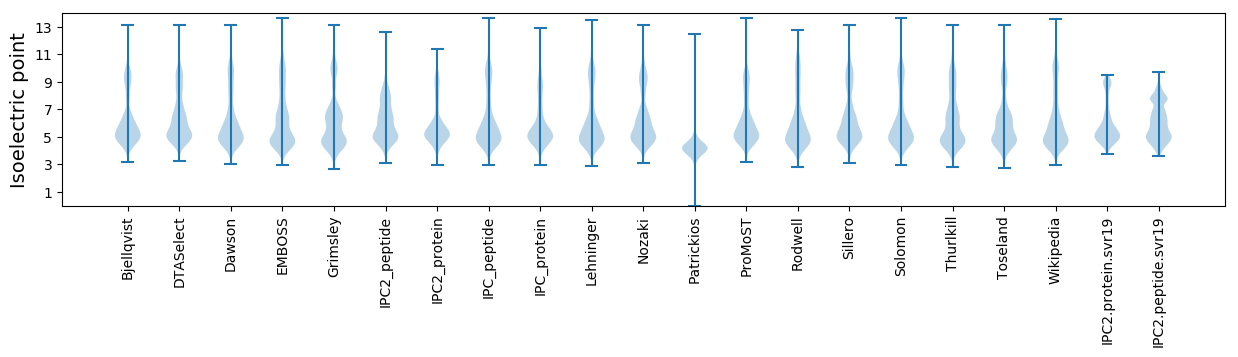
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T1D2N7|A0A2T1D2N7_9CYAN Thiazole synthase (Fragment) OS=filamentous cyanobacterium CCP2 OX=2107700 GN=C7B76_15640 PE=3 SV=1
MM1 pKa = 7.76PGNLRR6 pKa = 11.84ALKK9 pKa = 8.79EE10 pKa = 4.39TIATLQDD17 pKa = 3.22RR18 pKa = 11.84LHH20 pKa = 6.93PIVLPIYY27 pKa = 10.13CYY29 pKa = 10.17TEE31 pKa = 3.45ISKK34 pKa = 10.29VDD36 pKa = 3.59EE37 pKa = 4.53FLTPQLFTPGSDD49 pKa = 2.51IAIGFRR55 pKa = 11.84PSQIVWGRR63 pKa = 11.84TGNDD67 pKa = 2.91AVLGYY72 pKa = 9.53QPPASQTVQPQLDD85 pKa = 3.76ILIGDD90 pKa = 4.81LAIEE94 pKa = 4.72DD95 pKa = 3.7PAFRR99 pKa = 11.84QWSDD103 pKa = 3.09TFILGDD109 pKa = 3.39WTRR112 pKa = 11.84PYY114 pKa = 10.92YY115 pKa = 9.92IDD117 pKa = 4.75PEE119 pKa = 4.51PGFLGLNNFAILADD133 pKa = 4.55FDD135 pKa = 4.24PTLDD139 pKa = 3.92TIQLFGSANDD149 pKa = 3.74YY150 pKa = 11.07QLLDD154 pKa = 4.07LGFGTGIAFQTSTGPDD170 pKa = 3.57LIGFALGASGLNLAEE185 pKa = 5.44DD186 pKa = 4.02YY187 pKa = 11.41FEE189 pKa = 5.36FRR191 pKa = 11.84GTTPPPGPEE200 pKa = 3.77VPQIQQFGTTEE211 pKa = 3.64FDD213 pKa = 3.14IPLSISSDD221 pKa = 3.39PFGNIYY227 pKa = 10.06VAGGTNASLIEE238 pKa = 4.27TNEE241 pKa = 3.73GLRR244 pKa = 11.84DD245 pKa = 3.9NFVTKK250 pKa = 10.74YY251 pKa = 10.47NSQGQEE257 pKa = 3.88LFSLQFGTEE266 pKa = 3.96GFEE269 pKa = 4.2TIYY272 pKa = 11.1GIDD275 pKa = 3.71TDD277 pKa = 4.58DD278 pKa = 4.82LGNFYY283 pKa = 9.33ITGVTDD289 pKa = 4.47GEE291 pKa = 4.41LASPRR296 pKa = 11.84QSALLDD302 pKa = 3.67TFVAKK307 pKa = 10.72YY308 pKa = 10.29DD309 pKa = 4.08GEE311 pKa = 5.41GNQQWIRR318 pKa = 11.84QIGEE322 pKa = 3.87NLIFNAFNIAVDD334 pKa = 4.33GPTGDD339 pKa = 3.73VFISGANVKK348 pKa = 9.9PDD350 pKa = 3.86LANPDD355 pKa = 3.59DD356 pKa = 4.37AFVIKK361 pKa = 10.42FDD363 pKa = 4.31TDD365 pKa = 4.16GNQQWFTEE373 pKa = 4.19TGTTGFLNFDD383 pKa = 3.45EE384 pKa = 5.18TYY386 pKa = 10.93GLTVANDD393 pKa = 3.65GSVYY397 pKa = 9.49ATGWTVGDD405 pKa = 4.07LGGPNQGLYY414 pKa = 10.89DD415 pKa = 3.71NWIAKK420 pKa = 9.51YY421 pKa = 10.93DD422 pKa = 3.68NTTGEE427 pKa = 4.57EE428 pKa = 4.28VWVVQYY434 pKa = 7.19GTPDD438 pKa = 3.8YY439 pKa = 10.2EE440 pKa = 4.24WSWDD444 pKa = 3.46VRR446 pKa = 11.84TDD448 pKa = 3.37SLGNVYY454 pKa = 8.59TAGWTLGTLGDD465 pKa = 3.86EE466 pKa = 4.64SFGSFDD472 pKa = 4.81AYY474 pKa = 9.03LTKK477 pKa = 10.45YY478 pKa = 10.78DD479 pKa = 4.27SDD481 pKa = 4.69GNPLWVQQFGTPEE494 pKa = 4.56DD495 pKa = 3.8DD496 pKa = 3.13QAYY499 pKa = 10.87SLFIDD504 pKa = 3.27PHH506 pKa = 8.02DD507 pKa = 4.59NIFVGGYY514 pKa = 4.91TQGDD518 pKa = 3.55FGGQNVGSFDD528 pKa = 2.93AWIAKK533 pKa = 9.51FNTEE537 pKa = 4.99GEE539 pKa = 4.6KK540 pKa = 10.02IWINQFGTPNRR551 pKa = 11.84DD552 pKa = 2.97EE553 pKa = 5.52LYY555 pKa = 11.26GLTADD560 pKa = 4.38NAGNLYY566 pKa = 9.43ATGITQGSLGEE577 pKa = 4.2LNAGSFDD584 pKa = 3.32GWTAKK589 pKa = 10.65LDD591 pKa = 3.72VATGQLLDD599 pKa = 4.11FSGQAIDD606 pKa = 4.66APVEE610 pKa = 4.07EE611 pKa = 5.41DD612 pKa = 3.94CDD614 pKa = 4.27PLTGMPINSEE624 pKa = 4.14DD625 pKa = 3.6AVHH628 pKa = 7.14PEE630 pKa = 3.78AEE632 pKa = 4.28QPQLSNVEE640 pKa = 3.82IAQANGMSTALKK652 pKa = 10.46DD653 pKa = 3.58STFTPDD659 pKa = 2.77ILTGGSEE666 pKa = 4.07PPIEE670 pKa = 4.87EE671 pKa = 3.83IANPSGIDD679 pKa = 3.21LGAFFAAGSNPYY691 pKa = 9.63QAVSIQSLFANAFGLEE707 pKa = 3.85GVMDD711 pKa = 4.4NMPSYY716 pKa = 10.56QAVSSSSAFSQSS728 pKa = 2.59
MM1 pKa = 7.76PGNLRR6 pKa = 11.84ALKK9 pKa = 8.79EE10 pKa = 4.39TIATLQDD17 pKa = 3.22RR18 pKa = 11.84LHH20 pKa = 6.93PIVLPIYY27 pKa = 10.13CYY29 pKa = 10.17TEE31 pKa = 3.45ISKK34 pKa = 10.29VDD36 pKa = 3.59EE37 pKa = 4.53FLTPQLFTPGSDD49 pKa = 2.51IAIGFRR55 pKa = 11.84PSQIVWGRR63 pKa = 11.84TGNDD67 pKa = 2.91AVLGYY72 pKa = 9.53QPPASQTVQPQLDD85 pKa = 3.76ILIGDD90 pKa = 4.81LAIEE94 pKa = 4.72DD95 pKa = 3.7PAFRR99 pKa = 11.84QWSDD103 pKa = 3.09TFILGDD109 pKa = 3.39WTRR112 pKa = 11.84PYY114 pKa = 10.92YY115 pKa = 9.92IDD117 pKa = 4.75PEE119 pKa = 4.51PGFLGLNNFAILADD133 pKa = 4.55FDD135 pKa = 4.24PTLDD139 pKa = 3.92TIQLFGSANDD149 pKa = 3.74YY150 pKa = 11.07QLLDD154 pKa = 4.07LGFGTGIAFQTSTGPDD170 pKa = 3.57LIGFALGASGLNLAEE185 pKa = 5.44DD186 pKa = 4.02YY187 pKa = 11.41FEE189 pKa = 5.36FRR191 pKa = 11.84GTTPPPGPEE200 pKa = 3.77VPQIQQFGTTEE211 pKa = 3.64FDD213 pKa = 3.14IPLSISSDD221 pKa = 3.39PFGNIYY227 pKa = 10.06VAGGTNASLIEE238 pKa = 4.27TNEE241 pKa = 3.73GLRR244 pKa = 11.84DD245 pKa = 3.9NFVTKK250 pKa = 10.74YY251 pKa = 10.47NSQGQEE257 pKa = 3.88LFSLQFGTEE266 pKa = 3.96GFEE269 pKa = 4.2TIYY272 pKa = 11.1GIDD275 pKa = 3.71TDD277 pKa = 4.58DD278 pKa = 4.82LGNFYY283 pKa = 9.33ITGVTDD289 pKa = 4.47GEE291 pKa = 4.41LASPRR296 pKa = 11.84QSALLDD302 pKa = 3.67TFVAKK307 pKa = 10.72YY308 pKa = 10.29DD309 pKa = 4.08GEE311 pKa = 5.41GNQQWIRR318 pKa = 11.84QIGEE322 pKa = 3.87NLIFNAFNIAVDD334 pKa = 4.33GPTGDD339 pKa = 3.73VFISGANVKK348 pKa = 9.9PDD350 pKa = 3.86LANPDD355 pKa = 3.59DD356 pKa = 4.37AFVIKK361 pKa = 10.42FDD363 pKa = 4.31TDD365 pKa = 4.16GNQQWFTEE373 pKa = 4.19TGTTGFLNFDD383 pKa = 3.45EE384 pKa = 5.18TYY386 pKa = 10.93GLTVANDD393 pKa = 3.65GSVYY397 pKa = 9.49ATGWTVGDD405 pKa = 4.07LGGPNQGLYY414 pKa = 10.89DD415 pKa = 3.71NWIAKK420 pKa = 9.51YY421 pKa = 10.93DD422 pKa = 3.68NTTGEE427 pKa = 4.57EE428 pKa = 4.28VWVVQYY434 pKa = 7.19GTPDD438 pKa = 3.8YY439 pKa = 10.2EE440 pKa = 4.24WSWDD444 pKa = 3.46VRR446 pKa = 11.84TDD448 pKa = 3.37SLGNVYY454 pKa = 8.59TAGWTLGTLGDD465 pKa = 3.86EE466 pKa = 4.64SFGSFDD472 pKa = 4.81AYY474 pKa = 9.03LTKK477 pKa = 10.45YY478 pKa = 10.78DD479 pKa = 4.27SDD481 pKa = 4.69GNPLWVQQFGTPEE494 pKa = 4.56DD495 pKa = 3.8DD496 pKa = 3.13QAYY499 pKa = 10.87SLFIDD504 pKa = 3.27PHH506 pKa = 8.02DD507 pKa = 4.59NIFVGGYY514 pKa = 4.91TQGDD518 pKa = 3.55FGGQNVGSFDD528 pKa = 2.93AWIAKK533 pKa = 9.51FNTEE537 pKa = 4.99GEE539 pKa = 4.6KK540 pKa = 10.02IWINQFGTPNRR551 pKa = 11.84DD552 pKa = 2.97EE553 pKa = 5.52LYY555 pKa = 11.26GLTADD560 pKa = 4.38NAGNLYY566 pKa = 9.43ATGITQGSLGEE577 pKa = 4.2LNAGSFDD584 pKa = 3.32GWTAKK589 pKa = 10.65LDD591 pKa = 3.72VATGQLLDD599 pKa = 4.11FSGQAIDD606 pKa = 4.66APVEE610 pKa = 4.07EE611 pKa = 5.41DD612 pKa = 3.94CDD614 pKa = 4.27PLTGMPINSEE624 pKa = 4.14DD625 pKa = 3.6AVHH628 pKa = 7.14PEE630 pKa = 3.78AEE632 pKa = 4.28QPQLSNVEE640 pKa = 3.82IAQANGMSTALKK652 pKa = 10.46DD653 pKa = 3.58STFTPDD659 pKa = 2.77ILTGGSEE666 pKa = 4.07PPIEE670 pKa = 4.87EE671 pKa = 3.83IANPSGIDD679 pKa = 3.21LGAFFAAGSNPYY691 pKa = 9.63QAVSIQSLFANAFGLEE707 pKa = 3.85GVMDD711 pKa = 4.4NMPSYY716 pKa = 10.56QAVSSSSAFSQSS728 pKa = 2.59
Molecular weight: 78.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T1CJJ5|A0A2T1CJJ5_9CYAN Cobalt transport protein CbiN OS=filamentous cyanobacterium CCP2 OX=2107700 GN=cbiN PE=3 SV=1
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 9.07RR13 pKa = 11.84KK14 pKa = 8.17RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 9.95NGRR29 pKa = 11.84RR30 pKa = 11.84VIKK33 pKa = 10.23ARR35 pKa = 11.84RR36 pKa = 11.84SRR38 pKa = 11.84GRR40 pKa = 11.84VRR42 pKa = 11.84LAVV45 pKa = 3.37
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 9.07RR13 pKa = 11.84KK14 pKa = 8.17RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 9.95NGRR29 pKa = 11.84RR30 pKa = 11.84VIKK33 pKa = 10.23ARR35 pKa = 11.84RR36 pKa = 11.84SRR38 pKa = 11.84GRR40 pKa = 11.84VRR42 pKa = 11.84LAVV45 pKa = 3.37
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1872383 |
27 |
2476 |
301.4 |
33.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.769 ± 0.034 | 0.965 ± 0.011 |
5.166 ± 0.023 | 6.33 ± 0.033 |
3.911 ± 0.02 | 6.849 ± 0.036 |
2.088 ± 0.018 | 6.029 ± 0.025 |
3.439 ± 0.03 | 11.081 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.003 ± 0.014 | 3.615 ± 0.023 |
5.111 ± 0.025 | 5.594 ± 0.031 |
5.816 ± 0.027 | 6.344 ± 0.025 |
5.658 ± 0.026 | 6.875 ± 0.026 |
1.512 ± 0.015 | 2.844 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
