
Penaeid shrimp infectious myonecrosis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
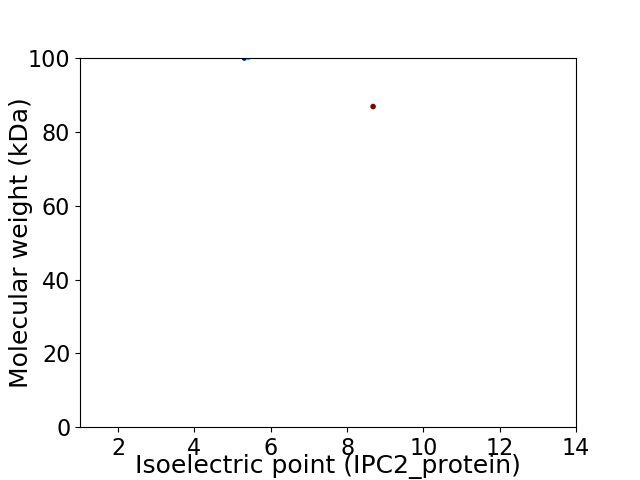
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q24M82|Q24M82_9VIRU Structural protein OS=Penaeid shrimp infectious myonecrosis virus OX=282786 PE=4 SV=1
MM1 pKa = 7.21HH2 pKa = 7.08VEE4 pKa = 3.79NGNIVSMEE12 pKa = 4.06NQSEE16 pKa = 4.08IDD18 pKa = 3.63SQTKK22 pKa = 9.54FFSLLEE28 pKa = 4.79DD29 pKa = 4.34DD30 pKa = 4.47NKK32 pKa = 11.16LPIVDD37 pKa = 4.26EE38 pKa = 4.4LRR40 pKa = 11.84VLADD44 pKa = 3.23MTAQRR49 pKa = 11.84SNVNTAGNHH58 pKa = 5.48LRR60 pKa = 11.84DD61 pKa = 3.88NDD63 pKa = 4.53SIRR66 pKa = 11.84ADD68 pKa = 2.92AVLANNTVRR77 pKa = 11.84NNCQIPIPVTTLIPRR92 pKa = 11.84QIRR95 pKa = 11.84GLNGVLVNQQLRR107 pKa = 11.84LQGIEE112 pKa = 3.74THH114 pKa = 6.1ITDD117 pKa = 3.87SYY119 pKa = 10.59ISKK122 pKa = 10.61AEE124 pKa = 3.81PSDD127 pKa = 3.73YY128 pKa = 11.0SKK130 pKa = 11.03QLSEE134 pKa = 4.18MVNAQKK140 pKa = 10.04TSTWRR145 pKa = 11.84ANNIASQGWDD155 pKa = 2.97MFDD158 pKa = 3.0TVQLNTNISQKK169 pKa = 10.59DD170 pKa = 3.63LSMDD174 pKa = 3.54TALTKK179 pKa = 10.93LMLLYY184 pKa = 10.68QLTTQNLPATQLPSSIYY201 pKa = 10.05SAFDD205 pKa = 3.06SRR207 pKa = 11.84TQPTLQDD214 pKa = 4.56GIWGINNGVNIFGEE228 pKa = 4.31QCGGLAAPVFPFSGGTGEE246 pKa = 4.19ITFHH250 pKa = 6.41LTLQSVPQEE259 pKa = 4.18FQEE262 pKa = 4.34SAIFVPATALQAAKK276 pKa = 10.32EE277 pKa = 4.22GARR280 pKa = 11.84TLAMYY285 pKa = 11.21VLMFAEE291 pKa = 5.25WPFGMYY297 pKa = 9.06TKK299 pKa = 10.27TKK301 pKa = 8.87QTTDD305 pKa = 2.97NAGNNQSDD313 pKa = 3.65QIFIHH318 pKa = 6.17SEE320 pKa = 4.12STVHH324 pKa = 7.25IPGQKK329 pKa = 9.48QMHH332 pKa = 6.05IVLPRR337 pKa = 11.84KK338 pKa = 10.06VNMVNPTTIAEE349 pKa = 3.98ANARR353 pKa = 11.84VVIQPTYY360 pKa = 8.63GTVAAGAGVANGNINVAAVGVALPTVNLTDD390 pKa = 4.23YY391 pKa = 10.33LVSWATDD398 pKa = 3.58FTLGDD403 pKa = 3.2IKK405 pKa = 11.06QLVEE409 pKa = 4.12RR410 pKa = 11.84MKK412 pKa = 8.68TTLPISRR419 pKa = 11.84DD420 pKa = 3.09LMAARR425 pKa = 11.84QNAMLLSTLFPPLIQSNVASDD446 pKa = 3.59TKK448 pKa = 10.45EE449 pKa = 4.03VPGTAGAYY457 pKa = 6.6TACLANLGIPEE468 pKa = 4.46TLTVNWGVDD477 pKa = 3.28INVQPLYY484 pKa = 10.78QLLEE488 pKa = 4.12TDD490 pKa = 3.11ITAHH494 pKa = 5.74NRR496 pKa = 11.84YY497 pKa = 9.11VLNLFKK503 pKa = 10.76RR504 pKa = 11.84EE505 pKa = 3.88EE506 pKa = 4.38VVAGAYY512 pKa = 8.82EE513 pKa = 4.39FGWLGHH519 pKa = 5.57MASYY523 pKa = 10.81MMGLLLTMNISSVFNVWYY541 pKa = 7.35STRR544 pKa = 11.84RR545 pKa = 11.84ISTKK549 pKa = 10.58AWDD552 pKa = 3.45TAYY555 pKa = 10.79DD556 pKa = 3.98SNIQAYY562 pKa = 10.05QDD564 pKa = 2.88MHH566 pKa = 7.63YY567 pKa = 11.24QMFSWSSMQGSIAPAMVDD585 pKa = 3.58EE586 pKa = 4.97ILHH589 pKa = 5.58NLCGQMFGFSLPLRR603 pKa = 11.84QVLFNALPITFSSFGSWMLPRR624 pKa = 11.84VSDD627 pKa = 3.79GFQTVRR633 pKa = 11.84YY634 pKa = 8.84YY635 pKa = 11.11DD636 pKa = 3.3VGPPVINAKK645 pKa = 9.98RR646 pKa = 11.84DD647 pKa = 3.57GEE649 pKa = 4.71VPVSMIDD656 pKa = 2.56AWTYY660 pKa = 11.24KK661 pKa = 9.42FTEE664 pKa = 4.27KK665 pKa = 10.51LPKK668 pKa = 10.28SFLPWPMPEE677 pKa = 4.64GKK679 pKa = 9.76DD680 pKa = 3.19STMGYY685 pKa = 10.46DD686 pKa = 3.71PEE688 pKa = 5.43KK689 pKa = 10.85EE690 pKa = 3.68PALIDD695 pKa = 3.61NSNEE699 pKa = 3.73TGNVFRR705 pKa = 11.84PFMARR710 pKa = 11.84NGNNSNYY717 pKa = 10.19LPTNYY722 pKa = 10.08TIDD725 pKa = 3.52VSQNGHH731 pKa = 6.86DD732 pKa = 3.73EE733 pKa = 4.12SCINVDD739 pKa = 4.55LFNNVAGVTLTNYY752 pKa = 10.74DD753 pKa = 3.48GTATNADD760 pKa = 3.75VVPTGSYY767 pKa = 9.59IKK769 pKa = 10.27QRR771 pKa = 11.84AMPINANAVRR781 pKa = 11.84PTEE784 pKa = 4.02TLDD787 pKa = 3.68AANHH791 pKa = 5.42TKK793 pKa = 10.33PFAIEE798 pKa = 3.96GGRR801 pKa = 11.84LVYY804 pKa = 10.78LGGTIANTTNVVNAMQRR821 pKa = 11.84KK822 pKa = 8.09QRR824 pKa = 11.84LSKK827 pKa = 10.34PAFKK831 pKa = 9.46WAHH834 pKa = 5.1AQRR837 pKa = 11.84QRR839 pKa = 11.84VYY841 pKa = 10.69DD842 pKa = 3.61SSRR845 pKa = 11.84PGMDD849 pKa = 5.52AITKK853 pKa = 9.57LCARR857 pKa = 11.84KK858 pKa = 9.99SGFMNARR865 pKa = 11.84STAMMAPKK873 pKa = 9.55TGLSAVIDD881 pKa = 3.82QAPNTSQDD889 pKa = 4.11LIEE892 pKa = 4.66QPSQQEE898 pKa = 4.18VMDD901 pKa = 3.96MQATATVV908 pKa = 3.35
MM1 pKa = 7.21HH2 pKa = 7.08VEE4 pKa = 3.79NGNIVSMEE12 pKa = 4.06NQSEE16 pKa = 4.08IDD18 pKa = 3.63SQTKK22 pKa = 9.54FFSLLEE28 pKa = 4.79DD29 pKa = 4.34DD30 pKa = 4.47NKK32 pKa = 11.16LPIVDD37 pKa = 4.26EE38 pKa = 4.4LRR40 pKa = 11.84VLADD44 pKa = 3.23MTAQRR49 pKa = 11.84SNVNTAGNHH58 pKa = 5.48LRR60 pKa = 11.84DD61 pKa = 3.88NDD63 pKa = 4.53SIRR66 pKa = 11.84ADD68 pKa = 2.92AVLANNTVRR77 pKa = 11.84NNCQIPIPVTTLIPRR92 pKa = 11.84QIRR95 pKa = 11.84GLNGVLVNQQLRR107 pKa = 11.84LQGIEE112 pKa = 3.74THH114 pKa = 6.1ITDD117 pKa = 3.87SYY119 pKa = 10.59ISKK122 pKa = 10.61AEE124 pKa = 3.81PSDD127 pKa = 3.73YY128 pKa = 11.0SKK130 pKa = 11.03QLSEE134 pKa = 4.18MVNAQKK140 pKa = 10.04TSTWRR145 pKa = 11.84ANNIASQGWDD155 pKa = 2.97MFDD158 pKa = 3.0TVQLNTNISQKK169 pKa = 10.59DD170 pKa = 3.63LSMDD174 pKa = 3.54TALTKK179 pKa = 10.93LMLLYY184 pKa = 10.68QLTTQNLPATQLPSSIYY201 pKa = 10.05SAFDD205 pKa = 3.06SRR207 pKa = 11.84TQPTLQDD214 pKa = 4.56GIWGINNGVNIFGEE228 pKa = 4.31QCGGLAAPVFPFSGGTGEE246 pKa = 4.19ITFHH250 pKa = 6.41LTLQSVPQEE259 pKa = 4.18FQEE262 pKa = 4.34SAIFVPATALQAAKK276 pKa = 10.32EE277 pKa = 4.22GARR280 pKa = 11.84TLAMYY285 pKa = 11.21VLMFAEE291 pKa = 5.25WPFGMYY297 pKa = 9.06TKK299 pKa = 10.27TKK301 pKa = 8.87QTTDD305 pKa = 2.97NAGNNQSDD313 pKa = 3.65QIFIHH318 pKa = 6.17SEE320 pKa = 4.12STVHH324 pKa = 7.25IPGQKK329 pKa = 9.48QMHH332 pKa = 6.05IVLPRR337 pKa = 11.84KK338 pKa = 10.06VNMVNPTTIAEE349 pKa = 3.98ANARR353 pKa = 11.84VVIQPTYY360 pKa = 8.63GTVAAGAGVANGNINVAAVGVALPTVNLTDD390 pKa = 4.23YY391 pKa = 10.33LVSWATDD398 pKa = 3.58FTLGDD403 pKa = 3.2IKK405 pKa = 11.06QLVEE409 pKa = 4.12RR410 pKa = 11.84MKK412 pKa = 8.68TTLPISRR419 pKa = 11.84DD420 pKa = 3.09LMAARR425 pKa = 11.84QNAMLLSTLFPPLIQSNVASDD446 pKa = 3.59TKK448 pKa = 10.45EE449 pKa = 4.03VPGTAGAYY457 pKa = 6.6TACLANLGIPEE468 pKa = 4.46TLTVNWGVDD477 pKa = 3.28INVQPLYY484 pKa = 10.78QLLEE488 pKa = 4.12TDD490 pKa = 3.11ITAHH494 pKa = 5.74NRR496 pKa = 11.84YY497 pKa = 9.11VLNLFKK503 pKa = 10.76RR504 pKa = 11.84EE505 pKa = 3.88EE506 pKa = 4.38VVAGAYY512 pKa = 8.82EE513 pKa = 4.39FGWLGHH519 pKa = 5.57MASYY523 pKa = 10.81MMGLLLTMNISSVFNVWYY541 pKa = 7.35STRR544 pKa = 11.84RR545 pKa = 11.84ISTKK549 pKa = 10.58AWDD552 pKa = 3.45TAYY555 pKa = 10.79DD556 pKa = 3.98SNIQAYY562 pKa = 10.05QDD564 pKa = 2.88MHH566 pKa = 7.63YY567 pKa = 11.24QMFSWSSMQGSIAPAMVDD585 pKa = 3.58EE586 pKa = 4.97ILHH589 pKa = 5.58NLCGQMFGFSLPLRR603 pKa = 11.84QVLFNALPITFSSFGSWMLPRR624 pKa = 11.84VSDD627 pKa = 3.79GFQTVRR633 pKa = 11.84YY634 pKa = 8.84YY635 pKa = 11.11DD636 pKa = 3.3VGPPVINAKK645 pKa = 9.98RR646 pKa = 11.84DD647 pKa = 3.57GEE649 pKa = 4.71VPVSMIDD656 pKa = 2.56AWTYY660 pKa = 11.24KK661 pKa = 9.42FTEE664 pKa = 4.27KK665 pKa = 10.51LPKK668 pKa = 10.28SFLPWPMPEE677 pKa = 4.64GKK679 pKa = 9.76DD680 pKa = 3.19STMGYY685 pKa = 10.46DD686 pKa = 3.71PEE688 pKa = 5.43KK689 pKa = 10.85EE690 pKa = 3.68PALIDD695 pKa = 3.61NSNEE699 pKa = 3.73TGNVFRR705 pKa = 11.84PFMARR710 pKa = 11.84NGNNSNYY717 pKa = 10.19LPTNYY722 pKa = 10.08TIDD725 pKa = 3.52VSQNGHH731 pKa = 6.86DD732 pKa = 3.73EE733 pKa = 4.12SCINVDD739 pKa = 4.55LFNNVAGVTLTNYY752 pKa = 10.74DD753 pKa = 3.48GTATNADD760 pKa = 3.75VVPTGSYY767 pKa = 9.59IKK769 pKa = 10.27QRR771 pKa = 11.84AMPINANAVRR781 pKa = 11.84PTEE784 pKa = 4.02TLDD787 pKa = 3.68AANHH791 pKa = 5.42TKK793 pKa = 10.33PFAIEE798 pKa = 3.96GGRR801 pKa = 11.84LVYY804 pKa = 10.78LGGTIANTTNVVNAMQRR821 pKa = 11.84KK822 pKa = 8.09QRR824 pKa = 11.84LSKK827 pKa = 10.34PAFKK831 pKa = 9.46WAHH834 pKa = 5.1AQRR837 pKa = 11.84QRR839 pKa = 11.84VYY841 pKa = 10.69DD842 pKa = 3.61SSRR845 pKa = 11.84PGMDD849 pKa = 5.52AITKK853 pKa = 9.57LCARR857 pKa = 11.84KK858 pKa = 9.99SGFMNARR865 pKa = 11.84STAMMAPKK873 pKa = 9.55TGLSAVIDD881 pKa = 3.82QAPNTSQDD889 pKa = 4.11LIEE892 pKa = 4.66QPSQQEE898 pKa = 4.18VMDD901 pKa = 3.96MQATATVV908 pKa = 3.35
Molecular weight: 100.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q24M82|Q24M82_9VIRU Structural protein OS=Penaeid shrimp infectious myonecrosis virus OX=282786 PE=4 SV=1
MM1 pKa = 8.18DD2 pKa = 5.69EE3 pKa = 4.04IRR5 pKa = 11.84NYY7 pKa = 8.96THH9 pKa = 7.32LDD11 pKa = 3.5YY12 pKa = 11.25IFVQNICIYY21 pKa = 9.1MLVFGIDD28 pKa = 3.4TVKK31 pKa = 10.48HH32 pKa = 5.3FRR34 pKa = 11.84QIGLFNEE41 pKa = 3.88RR42 pKa = 11.84NEE44 pKa = 4.42FIEE47 pKa = 4.16IAKK50 pKa = 9.96QLSTKK55 pKa = 9.84GKK57 pKa = 10.13RR58 pKa = 11.84FVDD61 pKa = 3.92DD62 pKa = 3.63VDD64 pKa = 3.55NMKK67 pKa = 10.77QKK69 pKa = 9.97VCEE72 pKa = 3.56IATIVGYY79 pKa = 9.39MDD81 pKa = 4.86PNVDD85 pKa = 4.06KK86 pKa = 10.56IDD88 pKa = 3.46VMEE91 pKa = 4.54EE92 pKa = 4.15VNSLAAEE99 pKa = 4.38GNEE102 pKa = 4.27HH103 pKa = 7.47GIDD106 pKa = 3.33RR107 pKa = 11.84DD108 pKa = 3.28NWNDD112 pKa = 4.33LFTKK116 pKa = 8.76TCKK119 pKa = 10.25EE120 pKa = 3.6VMTWYY125 pKa = 10.14KK126 pKa = 9.88GHH128 pKa = 6.32EE129 pKa = 4.66FISFDD134 pKa = 3.95DD135 pKa = 4.37YY136 pKa = 11.35IKK138 pKa = 11.04EE139 pKa = 4.23GMWLTSGSSSIGKK152 pKa = 7.43VHH154 pKa = 5.05WTKK157 pKa = 11.03DD158 pKa = 3.32GEE160 pKa = 4.31NGKK163 pKa = 10.2FKK165 pKa = 10.87ARR167 pKa = 11.84KK168 pKa = 9.39NMLLQIYY175 pKa = 7.67TPQEE179 pKa = 3.67LANIVYY185 pKa = 9.94AWDD188 pKa = 3.91GKK190 pKa = 9.1LHH192 pKa = 5.97SRR194 pKa = 11.84VFIKK198 pKa = 11.08NEE200 pKa = 3.64MSKK203 pKa = 10.74LRR205 pKa = 11.84LAVASNIEE213 pKa = 4.12AYY215 pKa = 9.57IHH217 pKa = 5.95EE218 pKa = 4.82SYY220 pKa = 10.8MLFLYY225 pKa = 10.2GHH227 pKa = 6.35GFKK230 pKa = 10.44EE231 pKa = 4.4YY232 pKa = 10.66FGVTLDD238 pKa = 3.99EE239 pKa = 5.7KK240 pKa = 10.82PDD242 pKa = 3.51QQHH245 pKa = 4.28QRR247 pKa = 11.84EE248 pKa = 4.02IEE250 pKa = 4.18MIEE253 pKa = 4.1KK254 pKa = 10.09LQAGYY259 pKa = 10.45FGLPFDD265 pKa = 4.37YY266 pKa = 10.95ASFDD270 pKa = 3.59HH271 pKa = 6.68QPTTFEE277 pKa = 4.0VKK279 pKa = 9.62TMVRR283 pKa = 11.84RR284 pKa = 11.84VGEE287 pKa = 4.07IVVSQVPKK295 pKa = 9.84NYY297 pKa = 10.37YY298 pKa = 9.77YY299 pKa = 9.29QTQLLVNKK307 pKa = 9.81IVNAYY312 pKa = 9.35DD313 pKa = 3.3KK314 pKa = 10.96SYY316 pKa = 11.44LSGNIKK322 pKa = 8.18NTKK325 pKa = 9.62FEE327 pKa = 4.46NIKK330 pKa = 10.73VKK332 pKa = 10.73GGVPSGVRR340 pKa = 11.84ITSLLGNMWNAIITKK355 pKa = 9.84IAINNVIGIIGYY367 pKa = 9.77DD368 pKa = 3.91PISQISLRR376 pKa = 11.84GDD378 pKa = 3.3DD379 pKa = 3.83VAILSKK385 pKa = 11.1DD386 pKa = 3.24PAALYY391 pKa = 10.44LLRR394 pKa = 11.84LSYY397 pKa = 10.88AAINAIGKK405 pKa = 9.48DD406 pKa = 3.52SKK408 pKa = 11.22LGISPKK414 pKa = 9.65VCEE417 pKa = 4.14FLRR420 pKa = 11.84NEE422 pKa = 3.75ISVTGVRR429 pKa = 11.84GWTCRR434 pKa = 11.84GIGGISQRR442 pKa = 11.84KK443 pKa = 6.88PWNPQPWSPNDD454 pKa = 3.54EE455 pKa = 4.44VEE457 pKa = 4.58TNASNISLLEE467 pKa = 3.7RR468 pKa = 11.84RR469 pKa = 11.84AGIEE473 pKa = 3.9LQQLHH478 pKa = 7.19HH479 pKa = 6.51INKK482 pKa = 8.91VKK484 pKa = 10.03WSRR487 pKa = 11.84HH488 pKa = 3.13VRR490 pKa = 11.84QSYY493 pKa = 10.22KK494 pKa = 10.6YY495 pKa = 9.98LEE497 pKa = 4.17LPKK500 pKa = 10.39RR501 pKa = 11.84LGGFGIYY508 pKa = 10.01RR509 pKa = 11.84FQGWLPNGKK518 pKa = 9.54LPLAKK523 pKa = 10.09KK524 pKa = 9.86PLVNVEE530 pKa = 5.06DD531 pKa = 3.48IHH533 pKa = 6.89PSQEE537 pKa = 3.73LFLPLSEE544 pKa = 4.11QQKK547 pKa = 10.69KK548 pKa = 9.94ILAQVEE554 pKa = 4.66MTNKK558 pKa = 8.73MQTDD562 pKa = 5.34DD563 pKa = 3.86IPGTQKK569 pKa = 10.79LFSKK573 pKa = 10.29EE574 pKa = 3.59WIQKK578 pKa = 8.87VRR580 pKa = 11.84AKK582 pKa = 10.54KK583 pKa = 10.31IIWSRR588 pKa = 11.84NQTIPIHH595 pKa = 6.05TDD597 pKa = 2.49HH598 pKa = 6.81TVRR601 pKa = 11.84IPRR604 pKa = 11.84WDD606 pKa = 3.65EE607 pKa = 3.9KK608 pKa = 11.11IKK610 pKa = 10.25FPRR613 pKa = 11.84YY614 pKa = 7.93KK615 pKa = 10.25SEE617 pKa = 4.28YY618 pKa = 9.58ILNNKK623 pKa = 8.89INLTMEE629 pKa = 4.01QVLRR633 pKa = 11.84QYY635 pKa = 11.67NLLKK639 pKa = 10.5EE640 pKa = 4.18VEE642 pKa = 4.76RR643 pKa = 11.84YY644 pKa = 10.28DD645 pKa = 5.03KK646 pKa = 10.83DD647 pKa = 3.91LKK649 pKa = 10.96VPKK652 pKa = 10.42LLDD655 pKa = 3.73ILDD658 pKa = 3.45KK659 pKa = 10.31WFPVQSSKK667 pKa = 10.81IKK669 pKa = 9.19TYY671 pKa = 10.37EE672 pKa = 3.73SQGFHH677 pKa = 5.86RR678 pKa = 11.84TDD680 pKa = 4.5AINLAVGEE688 pKa = 4.69IPTEE692 pKa = 3.83PAVKK696 pKa = 10.12INPILINFVKK706 pKa = 10.56LHH708 pKa = 6.44LEE710 pKa = 3.98RR711 pKa = 11.84QGIRR715 pKa = 11.84HH716 pKa = 4.24QRR718 pKa = 11.84GRR720 pKa = 11.84NKK722 pKa = 9.43IAKK725 pKa = 9.89FIYY728 pKa = 10.03QKK730 pKa = 9.66TKK732 pKa = 9.28QAEE735 pKa = 4.02NMILQSSLQQMYY747 pKa = 10.24RR748 pKa = 11.84YY749 pKa = 9.9
MM1 pKa = 8.18DD2 pKa = 5.69EE3 pKa = 4.04IRR5 pKa = 11.84NYY7 pKa = 8.96THH9 pKa = 7.32LDD11 pKa = 3.5YY12 pKa = 11.25IFVQNICIYY21 pKa = 9.1MLVFGIDD28 pKa = 3.4TVKK31 pKa = 10.48HH32 pKa = 5.3FRR34 pKa = 11.84QIGLFNEE41 pKa = 3.88RR42 pKa = 11.84NEE44 pKa = 4.42FIEE47 pKa = 4.16IAKK50 pKa = 9.96QLSTKK55 pKa = 9.84GKK57 pKa = 10.13RR58 pKa = 11.84FVDD61 pKa = 3.92DD62 pKa = 3.63VDD64 pKa = 3.55NMKK67 pKa = 10.77QKK69 pKa = 9.97VCEE72 pKa = 3.56IATIVGYY79 pKa = 9.39MDD81 pKa = 4.86PNVDD85 pKa = 4.06KK86 pKa = 10.56IDD88 pKa = 3.46VMEE91 pKa = 4.54EE92 pKa = 4.15VNSLAAEE99 pKa = 4.38GNEE102 pKa = 4.27HH103 pKa = 7.47GIDD106 pKa = 3.33RR107 pKa = 11.84DD108 pKa = 3.28NWNDD112 pKa = 4.33LFTKK116 pKa = 8.76TCKK119 pKa = 10.25EE120 pKa = 3.6VMTWYY125 pKa = 10.14KK126 pKa = 9.88GHH128 pKa = 6.32EE129 pKa = 4.66FISFDD134 pKa = 3.95DD135 pKa = 4.37YY136 pKa = 11.35IKK138 pKa = 11.04EE139 pKa = 4.23GMWLTSGSSSIGKK152 pKa = 7.43VHH154 pKa = 5.05WTKK157 pKa = 11.03DD158 pKa = 3.32GEE160 pKa = 4.31NGKK163 pKa = 10.2FKK165 pKa = 10.87ARR167 pKa = 11.84KK168 pKa = 9.39NMLLQIYY175 pKa = 7.67TPQEE179 pKa = 3.67LANIVYY185 pKa = 9.94AWDD188 pKa = 3.91GKK190 pKa = 9.1LHH192 pKa = 5.97SRR194 pKa = 11.84VFIKK198 pKa = 11.08NEE200 pKa = 3.64MSKK203 pKa = 10.74LRR205 pKa = 11.84LAVASNIEE213 pKa = 4.12AYY215 pKa = 9.57IHH217 pKa = 5.95EE218 pKa = 4.82SYY220 pKa = 10.8MLFLYY225 pKa = 10.2GHH227 pKa = 6.35GFKK230 pKa = 10.44EE231 pKa = 4.4YY232 pKa = 10.66FGVTLDD238 pKa = 3.99EE239 pKa = 5.7KK240 pKa = 10.82PDD242 pKa = 3.51QQHH245 pKa = 4.28QRR247 pKa = 11.84EE248 pKa = 4.02IEE250 pKa = 4.18MIEE253 pKa = 4.1KK254 pKa = 10.09LQAGYY259 pKa = 10.45FGLPFDD265 pKa = 4.37YY266 pKa = 10.95ASFDD270 pKa = 3.59HH271 pKa = 6.68QPTTFEE277 pKa = 4.0VKK279 pKa = 9.62TMVRR283 pKa = 11.84RR284 pKa = 11.84VGEE287 pKa = 4.07IVVSQVPKK295 pKa = 9.84NYY297 pKa = 10.37YY298 pKa = 9.77YY299 pKa = 9.29QTQLLVNKK307 pKa = 9.81IVNAYY312 pKa = 9.35DD313 pKa = 3.3KK314 pKa = 10.96SYY316 pKa = 11.44LSGNIKK322 pKa = 8.18NTKK325 pKa = 9.62FEE327 pKa = 4.46NIKK330 pKa = 10.73VKK332 pKa = 10.73GGVPSGVRR340 pKa = 11.84ITSLLGNMWNAIITKK355 pKa = 9.84IAINNVIGIIGYY367 pKa = 9.77DD368 pKa = 3.91PISQISLRR376 pKa = 11.84GDD378 pKa = 3.3DD379 pKa = 3.83VAILSKK385 pKa = 11.1DD386 pKa = 3.24PAALYY391 pKa = 10.44LLRR394 pKa = 11.84LSYY397 pKa = 10.88AAINAIGKK405 pKa = 9.48DD406 pKa = 3.52SKK408 pKa = 11.22LGISPKK414 pKa = 9.65VCEE417 pKa = 4.14FLRR420 pKa = 11.84NEE422 pKa = 3.75ISVTGVRR429 pKa = 11.84GWTCRR434 pKa = 11.84GIGGISQRR442 pKa = 11.84KK443 pKa = 6.88PWNPQPWSPNDD454 pKa = 3.54EE455 pKa = 4.44VEE457 pKa = 4.58TNASNISLLEE467 pKa = 3.7RR468 pKa = 11.84RR469 pKa = 11.84AGIEE473 pKa = 3.9LQQLHH478 pKa = 7.19HH479 pKa = 6.51INKK482 pKa = 8.91VKK484 pKa = 10.03WSRR487 pKa = 11.84HH488 pKa = 3.13VRR490 pKa = 11.84QSYY493 pKa = 10.22KK494 pKa = 10.6YY495 pKa = 9.98LEE497 pKa = 4.17LPKK500 pKa = 10.39RR501 pKa = 11.84LGGFGIYY508 pKa = 10.01RR509 pKa = 11.84FQGWLPNGKK518 pKa = 9.54LPLAKK523 pKa = 10.09KK524 pKa = 9.86PLVNVEE530 pKa = 5.06DD531 pKa = 3.48IHH533 pKa = 6.89PSQEE537 pKa = 3.73LFLPLSEE544 pKa = 4.11QQKK547 pKa = 10.69KK548 pKa = 9.94ILAQVEE554 pKa = 4.66MTNKK558 pKa = 8.73MQTDD562 pKa = 5.34DD563 pKa = 3.86IPGTQKK569 pKa = 10.79LFSKK573 pKa = 10.29EE574 pKa = 3.59WIQKK578 pKa = 8.87VRR580 pKa = 11.84AKK582 pKa = 10.54KK583 pKa = 10.31IIWSRR588 pKa = 11.84NQTIPIHH595 pKa = 6.05TDD597 pKa = 2.49HH598 pKa = 6.81TVRR601 pKa = 11.84IPRR604 pKa = 11.84WDD606 pKa = 3.65EE607 pKa = 3.9KK608 pKa = 11.11IKK610 pKa = 10.25FPRR613 pKa = 11.84YY614 pKa = 7.93KK615 pKa = 10.25SEE617 pKa = 4.28YY618 pKa = 9.58ILNNKK623 pKa = 8.89INLTMEE629 pKa = 4.01QVLRR633 pKa = 11.84QYY635 pKa = 11.67NLLKK639 pKa = 10.5EE640 pKa = 4.18VEE642 pKa = 4.76RR643 pKa = 11.84YY644 pKa = 10.28DD645 pKa = 5.03KK646 pKa = 10.83DD647 pKa = 3.91LKK649 pKa = 10.96VPKK652 pKa = 10.42LLDD655 pKa = 3.73ILDD658 pKa = 3.45KK659 pKa = 10.31WFPVQSSKK667 pKa = 10.81IKK669 pKa = 9.19TYY671 pKa = 10.37EE672 pKa = 3.73SQGFHH677 pKa = 5.86RR678 pKa = 11.84TDD680 pKa = 4.5AINLAVGEE688 pKa = 4.69IPTEE692 pKa = 3.83PAVKK696 pKa = 10.12INPILINFVKK706 pKa = 10.56LHH708 pKa = 6.44LEE710 pKa = 3.98RR711 pKa = 11.84QGIRR715 pKa = 11.84HH716 pKa = 4.24QRR718 pKa = 11.84GRR720 pKa = 11.84NKK722 pKa = 9.43IAKK725 pKa = 9.89FIYY728 pKa = 10.03QKK730 pKa = 9.66TKK732 pKa = 9.28QAEE735 pKa = 4.02NMILQSSLQQMYY747 pKa = 10.24RR748 pKa = 11.84YY749 pKa = 9.9
Molecular weight: 86.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1657 |
749 |
908 |
828.5 |
93.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.699 ± 1.499 | 0.664 ± 0.002 |
5.069 ± 0.076 | 4.949 ± 0.777 |
3.742 ± 0.076 | 6.035 ± 0.016 |
1.992 ± 0.319 | 7.242 ± 1.232 |
5.975 ± 1.818 | 8.147 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.319 ± 0.536 | 6.82 ± 0.475 |
4.647 ± 0.454 | 5.613 ± 0.159 |
4.406 ± 0.313 | 6.095 ± 0.442 |
6.518 ± 1.315 | 6.699 ± 0.248 |
1.75 ± 0.148 | 3.621 ± 0.46 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
