
Hubei narna-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
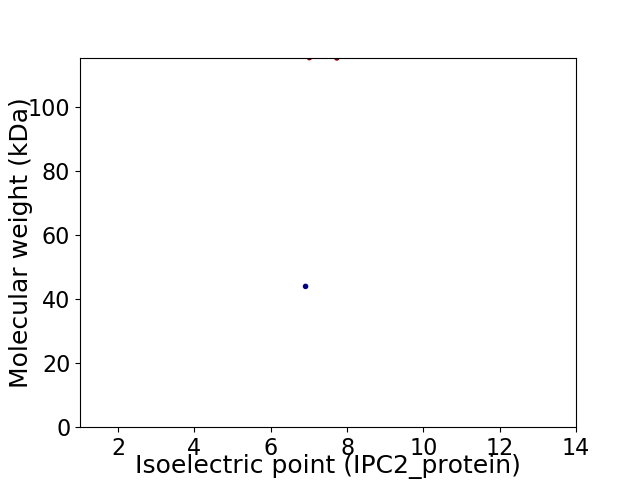
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
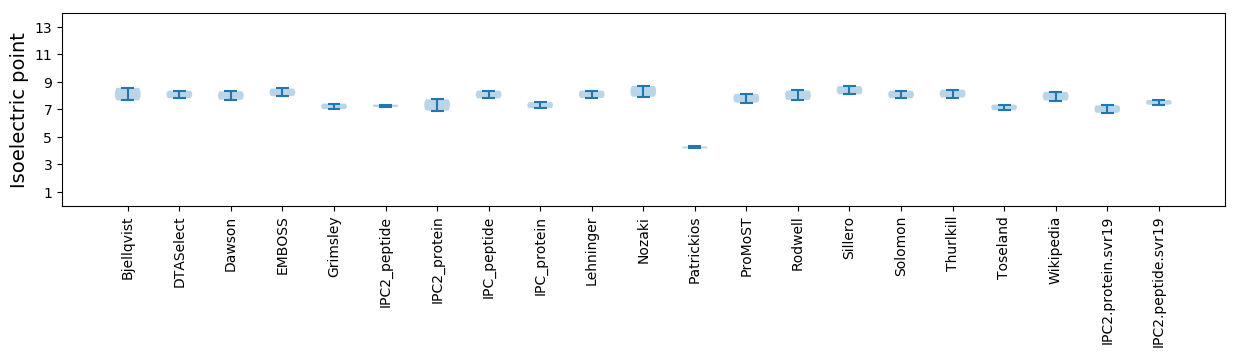
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIN5|A0A1L3KIN5_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 9 OX=1922962 PE=4 SV=1
MM1 pKa = 7.3PQAPQNNNKK10 pKa = 9.75QNTKK14 pKa = 9.97HH15 pKa = 6.63SNAAMQKK22 pKa = 9.97INEE25 pKa = 4.07QNKK28 pKa = 8.62NQNKK32 pKa = 7.84SQKK35 pKa = 9.69KK36 pKa = 7.67VNHH39 pKa = 5.83RR40 pKa = 11.84RR41 pKa = 11.84GYY43 pKa = 6.9QQRR46 pKa = 11.84PTLNQSQMFYY56 pKa = 11.03AALTCPFDD64 pKa = 4.48PNVLGVQVPDD74 pKa = 4.04PFPYY78 pKa = 8.33PTQVYY83 pKa = 9.06HH84 pKa = 5.16VHH86 pKa = 4.72QTTVIGNAANNSGSGAIAFLPNPVLSLIDD115 pKa = 3.2ITQANVGNITGSCIKK130 pKa = 9.43ATPFTRR136 pKa = 11.84YY137 pKa = 10.15GPLTPTNVSNAILGAITPTALSDD160 pKa = 3.3VFADD164 pKa = 3.53YY165 pKa = 10.79RR166 pKa = 11.84VVSWGIKK173 pKa = 9.34ISNLMPEE180 pKa = 4.71LVATGRR186 pKa = 11.84VIIAQIPLGDD196 pKa = 4.8TIPSYY201 pKa = 11.29PNLASAIQPVTLEE214 pKa = 4.48SIFGIDD220 pKa = 3.5PVFLGSSNILEE231 pKa = 4.76LPTGFQLTAQDD242 pKa = 4.33FLHH245 pKa = 7.28GDD247 pKa = 4.11LEE249 pKa = 4.76CGGMYY254 pKa = 10.15SAADD258 pKa = 3.47FWDD261 pKa = 4.46FKK263 pKa = 7.21TTRR266 pKa = 11.84DD267 pKa = 3.28IGKK270 pKa = 9.25LNVGGSPGTIFTGDD284 pKa = 3.53DD285 pKa = 3.55VAVTTSGLNYY295 pKa = 10.42GIGYY299 pKa = 10.14KK300 pKa = 10.8DD301 pKa = 3.26MTRR304 pKa = 11.84CRR306 pKa = 11.84GGSAIVIYY314 pKa = 10.07FEE316 pKa = 5.18GMPSNQIEE324 pKa = 4.24NFFQVEE330 pKa = 4.54TIYY333 pKa = 10.9HH334 pKa = 6.46LEE336 pKa = 4.0GTPNFSSISNNALISSTARR355 pKa = 11.84KK356 pKa = 6.98TAVGTTQNVEE366 pKa = 3.79QVMAKK371 pKa = 9.91ASKK374 pKa = 10.17VEE376 pKa = 4.93NVFTWIDD383 pKa = 3.25RR384 pKa = 11.84GADD387 pKa = 3.42FLNKK391 pKa = 10.0NKK393 pKa = 9.24STIMKK398 pKa = 9.87VGAAAMAFLL407 pKa = 4.85
MM1 pKa = 7.3PQAPQNNNKK10 pKa = 9.75QNTKK14 pKa = 9.97HH15 pKa = 6.63SNAAMQKK22 pKa = 9.97INEE25 pKa = 4.07QNKK28 pKa = 8.62NQNKK32 pKa = 7.84SQKK35 pKa = 9.69KK36 pKa = 7.67VNHH39 pKa = 5.83RR40 pKa = 11.84RR41 pKa = 11.84GYY43 pKa = 6.9QQRR46 pKa = 11.84PTLNQSQMFYY56 pKa = 11.03AALTCPFDD64 pKa = 4.48PNVLGVQVPDD74 pKa = 4.04PFPYY78 pKa = 8.33PTQVYY83 pKa = 9.06HH84 pKa = 5.16VHH86 pKa = 4.72QTTVIGNAANNSGSGAIAFLPNPVLSLIDD115 pKa = 3.2ITQANVGNITGSCIKK130 pKa = 9.43ATPFTRR136 pKa = 11.84YY137 pKa = 10.15GPLTPTNVSNAILGAITPTALSDD160 pKa = 3.3VFADD164 pKa = 3.53YY165 pKa = 10.79RR166 pKa = 11.84VVSWGIKK173 pKa = 9.34ISNLMPEE180 pKa = 4.71LVATGRR186 pKa = 11.84VIIAQIPLGDD196 pKa = 4.8TIPSYY201 pKa = 11.29PNLASAIQPVTLEE214 pKa = 4.48SIFGIDD220 pKa = 3.5PVFLGSSNILEE231 pKa = 4.76LPTGFQLTAQDD242 pKa = 4.33FLHH245 pKa = 7.28GDD247 pKa = 4.11LEE249 pKa = 4.76CGGMYY254 pKa = 10.15SAADD258 pKa = 3.47FWDD261 pKa = 4.46FKK263 pKa = 7.21TTRR266 pKa = 11.84DD267 pKa = 3.28IGKK270 pKa = 9.25LNVGGSPGTIFTGDD284 pKa = 3.53DD285 pKa = 3.55VAVTTSGLNYY295 pKa = 10.42GIGYY299 pKa = 10.14KK300 pKa = 10.8DD301 pKa = 3.26MTRR304 pKa = 11.84CRR306 pKa = 11.84GGSAIVIYY314 pKa = 10.07FEE316 pKa = 5.18GMPSNQIEE324 pKa = 4.24NFFQVEE330 pKa = 4.54TIYY333 pKa = 10.9HH334 pKa = 6.46LEE336 pKa = 4.0GTPNFSSISNNALISSTARR355 pKa = 11.84KK356 pKa = 6.98TAVGTTQNVEE366 pKa = 3.79QVMAKK371 pKa = 9.91ASKK374 pKa = 10.17VEE376 pKa = 4.93NVFTWIDD383 pKa = 3.25RR384 pKa = 11.84GADD387 pKa = 3.42FLNKK391 pKa = 10.0NKK393 pKa = 9.24STIMKK398 pKa = 9.87VGAAAMAFLL407 pKa = 4.85
Molecular weight: 43.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIN5|A0A1L3KIN5_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 9 OX=1922962 PE=4 SV=1
MM1 pKa = 7.82RR2 pKa = 11.84NHH4 pKa = 6.75LSFRR8 pKa = 11.84EE9 pKa = 4.14VCDD12 pKa = 3.52GCLGSLTRR20 pKa = 11.84GVPGTLKK27 pKa = 10.5NSKK30 pKa = 9.52VPRR33 pKa = 11.84KK34 pKa = 9.71RR35 pKa = 11.84YY36 pKa = 9.52FKK38 pKa = 10.66IPGVRR43 pKa = 11.84TWLKK47 pKa = 9.71TSGGNYY53 pKa = 7.29ITIFSTKK60 pKa = 10.06NKK62 pKa = 9.9IISKK66 pKa = 9.54LLKK69 pKa = 9.95EE70 pKa = 4.09AMKK73 pKa = 10.89VLTEE77 pKa = 3.72SHH79 pKa = 6.17GRR81 pKa = 11.84STTDD85 pKa = 3.88FSTFEE90 pKa = 3.4NWYY93 pKa = 7.8LHH95 pKa = 4.64EE96 pKa = 4.66VIRR99 pKa = 11.84IFKK102 pKa = 8.86ITDD105 pKa = 3.14KK106 pKa = 10.76TIEE109 pKa = 4.03PEE111 pKa = 3.9KK112 pKa = 10.61LFRR115 pKa = 11.84EE116 pKa = 4.4ATDD119 pKa = 3.68GFFEE123 pKa = 4.76KK124 pKa = 10.77ASFDD128 pKa = 4.99SNLEE132 pKa = 3.98WFWFNDD138 pKa = 3.15SYY140 pKa = 11.31LAEE143 pKa = 4.09ILQFVSAEE151 pKa = 3.75IHH153 pKa = 6.44ILTDD157 pKa = 3.69VIPDD161 pKa = 3.12FHH163 pKa = 8.12KK164 pKa = 10.53RR165 pKa = 11.84FSFEE169 pKa = 4.18EE170 pKa = 3.93QSKK173 pKa = 10.98LYY175 pKa = 10.88LNTLKK180 pKa = 10.38IPRR183 pKa = 11.84DD184 pKa = 3.59SFIKK188 pKa = 10.29HH189 pKa = 5.66IKK191 pKa = 9.56FHH193 pKa = 5.88TACPMASILDD203 pKa = 3.49QDD205 pKa = 4.92LPAKK209 pKa = 10.34PSDD212 pKa = 3.71FTGHH216 pKa = 5.09YY217 pKa = 9.49LIWTGSVKK225 pKa = 10.35RR226 pKa = 11.84YY227 pKa = 9.61LKK229 pKa = 10.78NILNRR234 pKa = 11.84RR235 pKa = 11.84NFNSKK240 pKa = 8.5DD241 pKa = 3.29TPLCLKK247 pKa = 10.23IGMGFLQGIKK257 pKa = 9.77RR258 pKa = 11.84GCATVPSSFLYY269 pKa = 10.74SEE271 pKa = 4.91VISHH275 pKa = 5.91IHH277 pKa = 6.86AMTTPPEE284 pKa = 4.13LVPGWEE290 pKa = 4.24YY291 pKa = 11.16TDD293 pKa = 4.37GFATKK298 pKa = 10.3YY299 pKa = 10.31SDD301 pKa = 5.86PILQTFGATCEE312 pKa = 4.22HH313 pKa = 6.64VLKK316 pKa = 9.23QTNKK320 pKa = 8.97VYY322 pKa = 10.59VPKK325 pKa = 10.17PYY327 pKa = 10.25EE328 pKa = 4.1PSHH331 pKa = 5.62NASFEE336 pKa = 4.03RR337 pKa = 11.84SRR339 pKa = 11.84GEE341 pKa = 3.64PEE343 pKa = 3.63FEE345 pKa = 3.56IEE347 pKa = 4.24KK348 pKa = 9.93FVKK351 pKa = 9.96GGAYY355 pKa = 9.09MEE357 pKa = 4.66VVHH360 pKa = 6.93SLGLQKK366 pKa = 9.11ITKK369 pKa = 9.22EE370 pKa = 4.21VILKK374 pKa = 10.56GDD376 pKa = 3.32VGFNISYY383 pKa = 8.46EE384 pKa = 4.21LPDD387 pKa = 3.52MDD389 pKa = 5.55EE390 pKa = 4.07VAEE393 pKa = 4.19LCRR396 pKa = 11.84IKK398 pKa = 10.98YY399 pKa = 8.67PSRR402 pKa = 11.84FKK404 pKa = 11.01FVDD407 pKa = 3.72DD408 pKa = 4.54DD409 pKa = 4.5PLFKK413 pKa = 10.57EE414 pKa = 4.28YY415 pKa = 10.52FPEE418 pKa = 4.4LFVSSPQLSRR428 pKa = 11.84EE429 pKa = 3.96LSDD432 pKa = 3.34TQVIPLCEE440 pKa = 3.87PLKK443 pKa = 10.7IRR445 pKa = 11.84VITKK449 pKa = 10.55GEE451 pKa = 3.96ALPAYY456 pKa = 9.46LAKK459 pKa = 9.92TLQKK463 pKa = 9.83TMKK466 pKa = 10.03SYY468 pKa = 11.12INRR471 pKa = 11.84FPSMILTTRR480 pKa = 11.84PLKK483 pKa = 10.75VDD485 pKa = 3.52DD486 pKa = 5.81FRR488 pKa = 11.84DD489 pKa = 3.06VWARR493 pKa = 11.84EE494 pKa = 3.87KK495 pKa = 10.83AIEE498 pKa = 4.15SQLGINLKK506 pKa = 10.21FCEE509 pKa = 4.69HH510 pKa = 6.39VSGDD514 pKa = 3.66YY515 pKa = 10.38KK516 pKa = 11.24AATDD520 pKa = 3.69KK521 pKa = 11.61LNINFTKK528 pKa = 10.88LIFEE532 pKa = 4.54RR533 pKa = 11.84FMVVLNIPQADD544 pKa = 3.65RR545 pKa = 11.84DD546 pKa = 4.01VYY548 pKa = 11.01RR549 pKa = 11.84SVLYY553 pKa = 8.17EE554 pKa = 3.84QRR556 pKa = 11.84VHH558 pKa = 6.54YY559 pKa = 9.82PFAAKK564 pKa = 10.41LCRR567 pKa = 11.84DD568 pKa = 3.42KK569 pKa = 11.22KK570 pKa = 11.05LNSLNCLHH578 pKa = 7.09KK579 pKa = 11.16ANIEE583 pKa = 3.98AKK585 pKa = 10.12KK586 pKa = 10.42LYY588 pKa = 9.87KK589 pKa = 10.24LEE591 pKa = 3.92YY592 pKa = 9.67AKK594 pKa = 10.09WVSEE598 pKa = 3.8GSNFLTEE605 pKa = 4.28PVLPPLEE612 pKa = 4.15FAVEE616 pKa = 4.06QKK618 pKa = 10.62NGQLMGSILSFPILCLANLICYY640 pKa = 9.64KK641 pKa = 10.44CALDD645 pKa = 3.95EE646 pKa = 5.19YY647 pKa = 11.0INIDD651 pKa = 3.06NKK653 pKa = 10.54LRR655 pKa = 11.84SKK657 pKa = 10.97KK658 pKa = 9.86FVNVFDD664 pKa = 5.25LPVLINGDD672 pKa = 3.92DD673 pKa = 3.2IYY675 pKa = 11.26FRR677 pKa = 11.84SNPVFYY683 pKa = 9.96EE684 pKa = 3.37IWKK687 pKa = 9.85KK688 pKa = 10.87YY689 pKa = 9.88IGIAGFVLSIGKK701 pKa = 9.91NYY703 pKa = 7.34VHH705 pKa = 7.3KK706 pKa = 10.85SIFTINSQCFRR717 pKa = 11.84YY718 pKa = 9.99NDD720 pKa = 3.63EE721 pKa = 4.06QDD723 pKa = 4.05SIDD726 pKa = 4.17EE727 pKa = 3.98ITYY730 pKa = 10.82LNVGLLIGQSKK741 pKa = 10.58SGILGEE747 pKa = 5.47KK748 pKa = 10.22LPTWDD753 pKa = 4.73LYY755 pKa = 11.91NKK757 pKa = 8.07VTVGSYY763 pKa = 11.12NKK765 pKa = 9.56VDD767 pKa = 2.99SHH769 pKa = 7.21NRR771 pKa = 11.84FFYY774 pKa = 9.23YY775 pKa = 10.38HH776 pKa = 7.35KK777 pKa = 10.84DD778 pKa = 3.35SIAQISKK785 pKa = 9.93NGNYY789 pKa = 10.48NLFLPKK795 pKa = 10.5LLGGLGFIRR804 pKa = 11.84PDD806 pKa = 3.18PAIPVKK812 pKa = 9.75ITRR815 pKa = 11.84FQAQLGTYY823 pKa = 7.11FHH825 pKa = 6.52NRR827 pKa = 11.84IIAAYY832 pKa = 6.51KK833 pKa = 8.66TPEE836 pKa = 4.24KK837 pKa = 9.55EE838 pKa = 3.86LHH840 pKa = 5.91MSQAHH845 pKa = 7.09LIDD848 pKa = 3.6EE849 pKa = 5.0HH850 pKa = 8.82SPDD853 pKa = 4.41VYY855 pKa = 10.57DD856 pKa = 4.33TYY858 pKa = 11.43LGDD861 pKa = 4.98QIFQFIKK868 pKa = 10.22KK869 pKa = 10.1DD870 pKa = 3.49EE871 pKa = 4.25EE872 pKa = 4.34VPEE875 pKa = 5.16GYY877 pKa = 10.69LPLNLSKK884 pKa = 10.24RR885 pKa = 11.84TEE887 pKa = 3.99HH888 pKa = 7.02LFVHH892 pKa = 6.88DD893 pKa = 4.18WGNFEE898 pKa = 4.53PKK900 pKa = 10.55LEE902 pKa = 3.95FRR904 pKa = 11.84SIDD907 pKa = 3.71SQTLKK912 pKa = 10.68HH913 pKa = 5.9FRR915 pKa = 11.84EE916 pKa = 4.49SVKK919 pKa = 10.34KK920 pKa = 10.0YY921 pKa = 10.46QGDD924 pKa = 3.77KK925 pKa = 10.3CWFGAEE931 pKa = 4.23SAISGNYY938 pKa = 9.07PYY940 pKa = 11.35VLVRR944 pKa = 11.84RR945 pKa = 11.84VLDD948 pKa = 3.79DD949 pKa = 3.61VEE951 pKa = 4.82TYY953 pKa = 10.41QEE955 pKa = 3.86NLRR958 pKa = 11.84EE959 pKa = 4.0VVYY962 pKa = 11.21NKK964 pKa = 9.95VLEE967 pKa = 4.46NIKK970 pKa = 10.78DD971 pKa = 3.69YY972 pKa = 11.66FEE974 pKa = 4.5VKK976 pKa = 10.1EE977 pKa = 4.12KK978 pKa = 10.94LKK980 pKa = 10.63PLIFNGNPQVVTDD993 pKa = 3.46VTTFF997 pKa = 3.03
MM1 pKa = 7.82RR2 pKa = 11.84NHH4 pKa = 6.75LSFRR8 pKa = 11.84EE9 pKa = 4.14VCDD12 pKa = 3.52GCLGSLTRR20 pKa = 11.84GVPGTLKK27 pKa = 10.5NSKK30 pKa = 9.52VPRR33 pKa = 11.84KK34 pKa = 9.71RR35 pKa = 11.84YY36 pKa = 9.52FKK38 pKa = 10.66IPGVRR43 pKa = 11.84TWLKK47 pKa = 9.71TSGGNYY53 pKa = 7.29ITIFSTKK60 pKa = 10.06NKK62 pKa = 9.9IISKK66 pKa = 9.54LLKK69 pKa = 9.95EE70 pKa = 4.09AMKK73 pKa = 10.89VLTEE77 pKa = 3.72SHH79 pKa = 6.17GRR81 pKa = 11.84STTDD85 pKa = 3.88FSTFEE90 pKa = 3.4NWYY93 pKa = 7.8LHH95 pKa = 4.64EE96 pKa = 4.66VIRR99 pKa = 11.84IFKK102 pKa = 8.86ITDD105 pKa = 3.14KK106 pKa = 10.76TIEE109 pKa = 4.03PEE111 pKa = 3.9KK112 pKa = 10.61LFRR115 pKa = 11.84EE116 pKa = 4.4ATDD119 pKa = 3.68GFFEE123 pKa = 4.76KK124 pKa = 10.77ASFDD128 pKa = 4.99SNLEE132 pKa = 3.98WFWFNDD138 pKa = 3.15SYY140 pKa = 11.31LAEE143 pKa = 4.09ILQFVSAEE151 pKa = 3.75IHH153 pKa = 6.44ILTDD157 pKa = 3.69VIPDD161 pKa = 3.12FHH163 pKa = 8.12KK164 pKa = 10.53RR165 pKa = 11.84FSFEE169 pKa = 4.18EE170 pKa = 3.93QSKK173 pKa = 10.98LYY175 pKa = 10.88LNTLKK180 pKa = 10.38IPRR183 pKa = 11.84DD184 pKa = 3.59SFIKK188 pKa = 10.29HH189 pKa = 5.66IKK191 pKa = 9.56FHH193 pKa = 5.88TACPMASILDD203 pKa = 3.49QDD205 pKa = 4.92LPAKK209 pKa = 10.34PSDD212 pKa = 3.71FTGHH216 pKa = 5.09YY217 pKa = 9.49LIWTGSVKK225 pKa = 10.35RR226 pKa = 11.84YY227 pKa = 9.61LKK229 pKa = 10.78NILNRR234 pKa = 11.84RR235 pKa = 11.84NFNSKK240 pKa = 8.5DD241 pKa = 3.29TPLCLKK247 pKa = 10.23IGMGFLQGIKK257 pKa = 9.77RR258 pKa = 11.84GCATVPSSFLYY269 pKa = 10.74SEE271 pKa = 4.91VISHH275 pKa = 5.91IHH277 pKa = 6.86AMTTPPEE284 pKa = 4.13LVPGWEE290 pKa = 4.24YY291 pKa = 11.16TDD293 pKa = 4.37GFATKK298 pKa = 10.3YY299 pKa = 10.31SDD301 pKa = 5.86PILQTFGATCEE312 pKa = 4.22HH313 pKa = 6.64VLKK316 pKa = 9.23QTNKK320 pKa = 8.97VYY322 pKa = 10.59VPKK325 pKa = 10.17PYY327 pKa = 10.25EE328 pKa = 4.1PSHH331 pKa = 5.62NASFEE336 pKa = 4.03RR337 pKa = 11.84SRR339 pKa = 11.84GEE341 pKa = 3.64PEE343 pKa = 3.63FEE345 pKa = 3.56IEE347 pKa = 4.24KK348 pKa = 9.93FVKK351 pKa = 9.96GGAYY355 pKa = 9.09MEE357 pKa = 4.66VVHH360 pKa = 6.93SLGLQKK366 pKa = 9.11ITKK369 pKa = 9.22EE370 pKa = 4.21VILKK374 pKa = 10.56GDD376 pKa = 3.32VGFNISYY383 pKa = 8.46EE384 pKa = 4.21LPDD387 pKa = 3.52MDD389 pKa = 5.55EE390 pKa = 4.07VAEE393 pKa = 4.19LCRR396 pKa = 11.84IKK398 pKa = 10.98YY399 pKa = 8.67PSRR402 pKa = 11.84FKK404 pKa = 11.01FVDD407 pKa = 3.72DD408 pKa = 4.54DD409 pKa = 4.5PLFKK413 pKa = 10.57EE414 pKa = 4.28YY415 pKa = 10.52FPEE418 pKa = 4.4LFVSSPQLSRR428 pKa = 11.84EE429 pKa = 3.96LSDD432 pKa = 3.34TQVIPLCEE440 pKa = 3.87PLKK443 pKa = 10.7IRR445 pKa = 11.84VITKK449 pKa = 10.55GEE451 pKa = 3.96ALPAYY456 pKa = 9.46LAKK459 pKa = 9.92TLQKK463 pKa = 9.83TMKK466 pKa = 10.03SYY468 pKa = 11.12INRR471 pKa = 11.84FPSMILTTRR480 pKa = 11.84PLKK483 pKa = 10.75VDD485 pKa = 3.52DD486 pKa = 5.81FRR488 pKa = 11.84DD489 pKa = 3.06VWARR493 pKa = 11.84EE494 pKa = 3.87KK495 pKa = 10.83AIEE498 pKa = 4.15SQLGINLKK506 pKa = 10.21FCEE509 pKa = 4.69HH510 pKa = 6.39VSGDD514 pKa = 3.66YY515 pKa = 10.38KK516 pKa = 11.24AATDD520 pKa = 3.69KK521 pKa = 11.61LNINFTKK528 pKa = 10.88LIFEE532 pKa = 4.54RR533 pKa = 11.84FMVVLNIPQADD544 pKa = 3.65RR545 pKa = 11.84DD546 pKa = 4.01VYY548 pKa = 11.01RR549 pKa = 11.84SVLYY553 pKa = 8.17EE554 pKa = 3.84QRR556 pKa = 11.84VHH558 pKa = 6.54YY559 pKa = 9.82PFAAKK564 pKa = 10.41LCRR567 pKa = 11.84DD568 pKa = 3.42KK569 pKa = 11.22KK570 pKa = 11.05LNSLNCLHH578 pKa = 7.09KK579 pKa = 11.16ANIEE583 pKa = 3.98AKK585 pKa = 10.12KK586 pKa = 10.42LYY588 pKa = 9.87KK589 pKa = 10.24LEE591 pKa = 3.92YY592 pKa = 9.67AKK594 pKa = 10.09WVSEE598 pKa = 3.8GSNFLTEE605 pKa = 4.28PVLPPLEE612 pKa = 4.15FAVEE616 pKa = 4.06QKK618 pKa = 10.62NGQLMGSILSFPILCLANLICYY640 pKa = 9.64KK641 pKa = 10.44CALDD645 pKa = 3.95EE646 pKa = 5.19YY647 pKa = 11.0INIDD651 pKa = 3.06NKK653 pKa = 10.54LRR655 pKa = 11.84SKK657 pKa = 10.97KK658 pKa = 9.86FVNVFDD664 pKa = 5.25LPVLINGDD672 pKa = 3.92DD673 pKa = 3.2IYY675 pKa = 11.26FRR677 pKa = 11.84SNPVFYY683 pKa = 9.96EE684 pKa = 3.37IWKK687 pKa = 9.85KK688 pKa = 10.87YY689 pKa = 9.88IGIAGFVLSIGKK701 pKa = 9.91NYY703 pKa = 7.34VHH705 pKa = 7.3KK706 pKa = 10.85SIFTINSQCFRR717 pKa = 11.84YY718 pKa = 9.99NDD720 pKa = 3.63EE721 pKa = 4.06QDD723 pKa = 4.05SIDD726 pKa = 4.17EE727 pKa = 3.98ITYY730 pKa = 10.82LNVGLLIGQSKK741 pKa = 10.58SGILGEE747 pKa = 5.47KK748 pKa = 10.22LPTWDD753 pKa = 4.73LYY755 pKa = 11.91NKK757 pKa = 8.07VTVGSYY763 pKa = 11.12NKK765 pKa = 9.56VDD767 pKa = 2.99SHH769 pKa = 7.21NRR771 pKa = 11.84FFYY774 pKa = 9.23YY775 pKa = 10.38HH776 pKa = 7.35KK777 pKa = 10.84DD778 pKa = 3.35SIAQISKK785 pKa = 9.93NGNYY789 pKa = 10.48NLFLPKK795 pKa = 10.5LLGGLGFIRR804 pKa = 11.84PDD806 pKa = 3.18PAIPVKK812 pKa = 9.75ITRR815 pKa = 11.84FQAQLGTYY823 pKa = 7.11FHH825 pKa = 6.52NRR827 pKa = 11.84IIAAYY832 pKa = 6.51KK833 pKa = 8.66TPEE836 pKa = 4.24KK837 pKa = 9.55EE838 pKa = 3.86LHH840 pKa = 5.91MSQAHH845 pKa = 7.09LIDD848 pKa = 3.6EE849 pKa = 5.0HH850 pKa = 8.82SPDD853 pKa = 4.41VYY855 pKa = 10.57DD856 pKa = 4.33TYY858 pKa = 11.43LGDD861 pKa = 4.98QIFQFIKK868 pKa = 10.22KK869 pKa = 10.1DD870 pKa = 3.49EE871 pKa = 4.25EE872 pKa = 4.34VPEE875 pKa = 5.16GYY877 pKa = 10.69LPLNLSKK884 pKa = 10.24RR885 pKa = 11.84TEE887 pKa = 3.99HH888 pKa = 7.02LFVHH892 pKa = 6.88DD893 pKa = 4.18WGNFEE898 pKa = 4.53PKK900 pKa = 10.55LEE902 pKa = 3.95FRR904 pKa = 11.84SIDD907 pKa = 3.71SQTLKK912 pKa = 10.68HH913 pKa = 5.9FRR915 pKa = 11.84EE916 pKa = 4.49SVKK919 pKa = 10.34KK920 pKa = 10.0YY921 pKa = 10.46QGDD924 pKa = 3.77KK925 pKa = 10.3CWFGAEE931 pKa = 4.23SAISGNYY938 pKa = 9.07PYY940 pKa = 11.35VLVRR944 pKa = 11.84RR945 pKa = 11.84VLDD948 pKa = 3.79DD949 pKa = 3.61VEE951 pKa = 4.82TYY953 pKa = 10.41QEE955 pKa = 3.86NLRR958 pKa = 11.84EE959 pKa = 4.0VVYY962 pKa = 11.21NKK964 pKa = 9.95VLEE967 pKa = 4.46NIKK970 pKa = 10.78DD971 pKa = 3.69YY972 pKa = 11.66FEE974 pKa = 4.5VKK976 pKa = 10.1EE977 pKa = 4.12KK978 pKa = 10.94LKK980 pKa = 10.63PLIFNGNPQVVTDD993 pKa = 3.46VTTFF997 pKa = 3.03
Molecular weight: 115.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1404 |
407 |
997 |
702.0 |
79.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.271 ± 1.65 | 1.425 ± 0.236 |
5.057 ± 0.471 | 5.556 ± 1.527 |
6.125 ± 0.648 | 5.983 ± 1.137 |
2.279 ± 0.431 | 7.194 ± 0.095 |
7.55 ± 1.674 | 8.761 ± 1.27 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.567 ± 0.476 | 5.983 ± 1.269 |
5.342 ± 0.428 | 3.632 ± 1.08 |
3.846 ± 0.612 | 6.624 ± 0.137 |
6.054 ± 1.231 | 6.41 ± 0.251 |
1.068 ± 0.177 | 4.274 ± 0.709 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
