
Geodermatophilus sabuli
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Geodermatophilus
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
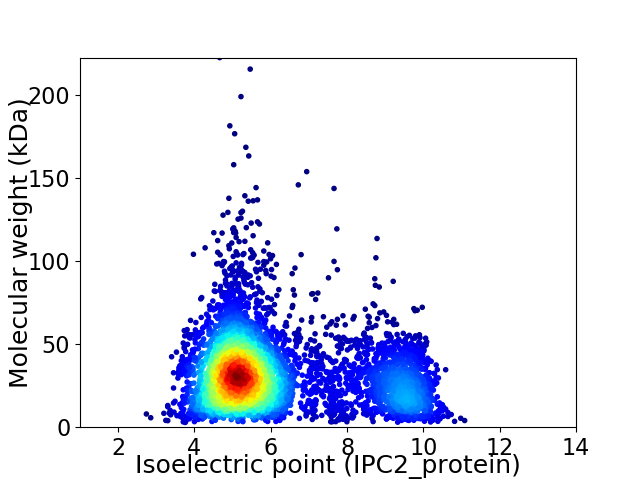
Virtual 2D-PAGE plot for 5204 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A285EFZ0|A0A285EFZ0_9ACTN Acyl-CoA thioesterase OS=Geodermatophilus sabuli OX=1564158 GN=SAMN06893097_105292 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 10.3LSKK5 pKa = 9.72RR6 pKa = 11.84TAALIATGLSGVMVLSACGGGDD28 pKa = 4.1DD29 pKa = 4.83EE30 pKa = 5.76GSSSADD36 pKa = 2.87GGSFSIYY43 pKa = 9.67IGEE46 pKa = 4.3PEE48 pKa = 4.27NPLMPGNTNEE58 pKa = 4.29TEE60 pKa = 4.22GGQVLDD66 pKa = 3.86SLFTGLVQYY75 pKa = 10.87DD76 pKa = 3.94PEE78 pKa = 4.34TNEE81 pKa = 3.74AAFTGVAEE89 pKa = 4.77SIEE92 pKa = 4.73SDD94 pKa = 4.19DD95 pKa = 3.61QTTWTVKK102 pKa = 10.86LNDD105 pKa = 2.97GWTFHH110 pKa = 7.46DD111 pKa = 5.08GSPVNAQSFVDD122 pKa = 3.58AWNYY126 pKa = 7.22TAYY129 pKa = 10.76SPNAQGNSYY138 pKa = 10.34FFANIVGYY146 pKa = 10.98GDD148 pKa = 3.7LQAPEE153 pKa = 4.64AGGDD157 pKa = 3.71PVATEE162 pKa = 3.88MSGLRR167 pKa = 11.84VVDD170 pKa = 4.26DD171 pKa = 3.66LTFEE175 pKa = 4.42VTLSSPYY182 pKa = 10.13AQWPTTVGYY191 pKa = 7.89TAFFPLPPAFFDD203 pKa = 4.09DD204 pKa = 4.05PAAFGEE210 pKa = 4.21QPIGNGPFQAVEE222 pKa = 4.13PFVPGQGVTLTRR234 pKa = 11.84YY235 pKa = 9.96EE236 pKa = 4.45DD237 pKa = 3.68FAGDD241 pKa = 3.71EE242 pKa = 4.1PAKK245 pKa = 10.71AEE247 pKa = 4.12SVEE250 pKa = 3.61YY251 pKa = 9.76RR252 pKa = 11.84VYY254 pKa = 11.0VEE256 pKa = 4.47QDD258 pKa = 2.75TAYY261 pKa = 9.82TDD263 pKa = 3.77LQGGSLDD270 pKa = 4.88IMDD273 pKa = 5.5TLPPDD278 pKa = 5.17AIASAEE284 pKa = 4.19SEE286 pKa = 3.99FGDD289 pKa = 4.44RR290 pKa = 11.84YY291 pKa = 10.57IEE293 pKa = 4.07TAQGDD298 pKa = 3.84ITSLGFPTYY307 pKa = 9.52DD308 pKa = 4.34QRR310 pKa = 11.84FADD313 pKa = 3.84PNVRR317 pKa = 11.84KK318 pKa = 9.59AFSMAIDD325 pKa = 3.7RR326 pKa = 11.84QAISDD331 pKa = 4.26AIFNGTRR338 pKa = 11.84TPATSFISPVVPGYY352 pKa = 10.64RR353 pKa = 11.84EE354 pKa = 4.25GACEE358 pKa = 3.62ACEE361 pKa = 4.41LNVEE365 pKa = 4.42EE366 pKa = 5.48ANSLLDD372 pKa = 3.3AAGFDD377 pKa = 3.54RR378 pKa = 11.84SQPVDD383 pKa = 2.84LWFNAGAGHH392 pKa = 6.64DD393 pKa = 3.49AWMEE397 pKa = 3.82AVGNQLRR404 pKa = 11.84EE405 pKa = 4.15NLGVEE410 pKa = 3.96YY411 pKa = 10.03QLRR414 pKa = 11.84GDD416 pKa = 4.32LPQAEE421 pKa = 4.58FLPLKK426 pKa = 9.57DD427 pKa = 3.92AKK429 pKa = 11.18GMTGPFRR436 pKa = 11.84DD437 pKa = 4.18AWIMDD442 pKa = 3.57YY443 pKa = 10.75PVAEE447 pKa = 4.34NFLGPLYY454 pKa = 10.76SSVALPPAGSNYY466 pKa = 9.09TFYY469 pKa = 11.36SNPQFDD475 pKa = 4.07QLLQQGNAAASDD487 pKa = 4.06DD488 pKa = 4.19EE489 pKa = 5.4AVAAYY494 pKa = 9.26QAAEE498 pKa = 4.27DD499 pKa = 4.12VLIADD504 pKa = 5.16LPSAPLFYY512 pKa = 10.72RR513 pKa = 11.84LNQGAHH519 pKa = 5.92SEE521 pKa = 4.17NVDD524 pKa = 3.37NVIIDD529 pKa = 3.34AFGRR533 pKa = 11.84IDD535 pKa = 3.23AAAVEE540 pKa = 4.93VVSS543 pKa = 4.44
MM1 pKa = 7.43KK2 pKa = 10.3LSKK5 pKa = 9.72RR6 pKa = 11.84TAALIATGLSGVMVLSACGGGDD28 pKa = 4.1DD29 pKa = 4.83EE30 pKa = 5.76GSSSADD36 pKa = 2.87GGSFSIYY43 pKa = 9.67IGEE46 pKa = 4.3PEE48 pKa = 4.27NPLMPGNTNEE58 pKa = 4.29TEE60 pKa = 4.22GGQVLDD66 pKa = 3.86SLFTGLVQYY75 pKa = 10.87DD76 pKa = 3.94PEE78 pKa = 4.34TNEE81 pKa = 3.74AAFTGVAEE89 pKa = 4.77SIEE92 pKa = 4.73SDD94 pKa = 4.19DD95 pKa = 3.61QTTWTVKK102 pKa = 10.86LNDD105 pKa = 2.97GWTFHH110 pKa = 7.46DD111 pKa = 5.08GSPVNAQSFVDD122 pKa = 3.58AWNYY126 pKa = 7.22TAYY129 pKa = 10.76SPNAQGNSYY138 pKa = 10.34FFANIVGYY146 pKa = 10.98GDD148 pKa = 3.7LQAPEE153 pKa = 4.64AGGDD157 pKa = 3.71PVATEE162 pKa = 3.88MSGLRR167 pKa = 11.84VVDD170 pKa = 4.26DD171 pKa = 3.66LTFEE175 pKa = 4.42VTLSSPYY182 pKa = 10.13AQWPTTVGYY191 pKa = 7.89TAFFPLPPAFFDD203 pKa = 4.09DD204 pKa = 4.05PAAFGEE210 pKa = 4.21QPIGNGPFQAVEE222 pKa = 4.13PFVPGQGVTLTRR234 pKa = 11.84YY235 pKa = 9.96EE236 pKa = 4.45DD237 pKa = 3.68FAGDD241 pKa = 3.71EE242 pKa = 4.1PAKK245 pKa = 10.71AEE247 pKa = 4.12SVEE250 pKa = 3.61YY251 pKa = 9.76RR252 pKa = 11.84VYY254 pKa = 11.0VEE256 pKa = 4.47QDD258 pKa = 2.75TAYY261 pKa = 9.82TDD263 pKa = 3.77LQGGSLDD270 pKa = 4.88IMDD273 pKa = 5.5TLPPDD278 pKa = 5.17AIASAEE284 pKa = 4.19SEE286 pKa = 3.99FGDD289 pKa = 4.44RR290 pKa = 11.84YY291 pKa = 10.57IEE293 pKa = 4.07TAQGDD298 pKa = 3.84ITSLGFPTYY307 pKa = 9.52DD308 pKa = 4.34QRR310 pKa = 11.84FADD313 pKa = 3.84PNVRR317 pKa = 11.84KK318 pKa = 9.59AFSMAIDD325 pKa = 3.7RR326 pKa = 11.84QAISDD331 pKa = 4.26AIFNGTRR338 pKa = 11.84TPATSFISPVVPGYY352 pKa = 10.64RR353 pKa = 11.84EE354 pKa = 4.25GACEE358 pKa = 3.62ACEE361 pKa = 4.41LNVEE365 pKa = 4.42EE366 pKa = 5.48ANSLLDD372 pKa = 3.3AAGFDD377 pKa = 3.54RR378 pKa = 11.84SQPVDD383 pKa = 2.84LWFNAGAGHH392 pKa = 6.64DD393 pKa = 3.49AWMEE397 pKa = 3.82AVGNQLRR404 pKa = 11.84EE405 pKa = 4.15NLGVEE410 pKa = 3.96YY411 pKa = 10.03QLRR414 pKa = 11.84GDD416 pKa = 4.32LPQAEE421 pKa = 4.58FLPLKK426 pKa = 9.57DD427 pKa = 3.92AKK429 pKa = 11.18GMTGPFRR436 pKa = 11.84DD437 pKa = 4.18AWIMDD442 pKa = 3.57YY443 pKa = 10.75PVAEE447 pKa = 4.34NFLGPLYY454 pKa = 10.76SSVALPPAGSNYY466 pKa = 9.09TFYY469 pKa = 11.36SNPQFDD475 pKa = 4.07QLLQQGNAAASDD487 pKa = 4.06DD488 pKa = 4.19EE489 pKa = 5.4AVAAYY494 pKa = 9.26QAAEE498 pKa = 4.27DD499 pKa = 4.12VLIADD504 pKa = 5.16LPSAPLFYY512 pKa = 10.72RR513 pKa = 11.84LNQGAHH519 pKa = 5.92SEE521 pKa = 4.17NVDD524 pKa = 3.37NVIIDD529 pKa = 3.34AFGRR533 pKa = 11.84IDD535 pKa = 3.23AAAVEE540 pKa = 4.93VVSS543 pKa = 4.44
Molecular weight: 58.17 kDa
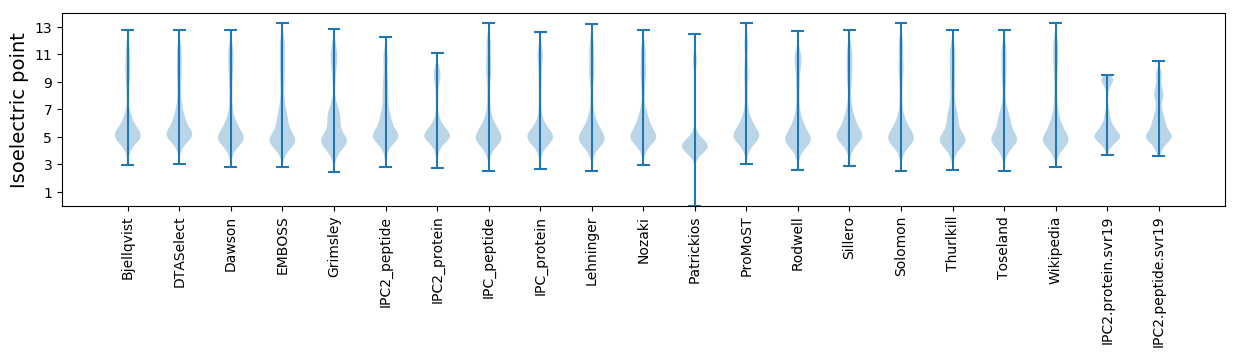
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A285E903|A0A285E903_9ACTN Drug resistance transporter EmrB/QacA subfamily OS=Geodermatophilus sabuli OX=1564158 GN=SAMN06893097_102205 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1639597 |
29 |
2138 |
315.1 |
33.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.326 ± 0.054 | 0.718 ± 0.009 |
6.187 ± 0.024 | 5.586 ± 0.029 |
2.611 ± 0.018 | 9.659 ± 0.03 |
2.076 ± 0.015 | 2.794 ± 0.022 |
1.265 ± 0.017 | 10.643 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.645 ± 0.013 | 1.433 ± 0.016 |
6.322 ± 0.031 | 2.686 ± 0.019 |
8.268 ± 0.037 | 4.758 ± 0.022 |
5.971 ± 0.025 | 9.81 ± 0.03 |
1.476 ± 0.014 | 1.766 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
