
Nostoc linckia NIES-25
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Nostocaceae; Nostoc; Nostoc linckia
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
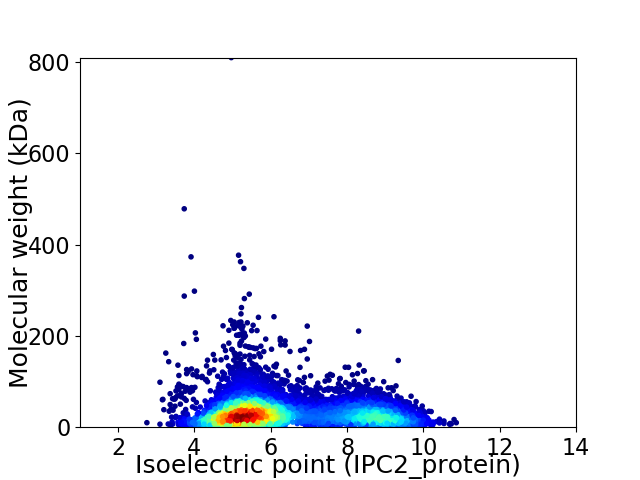
Virtual 2D-PAGE plot for 6881 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z4L9R1|A0A1Z4L9R1_NOSLI Uncharacterized protein OS=Nostoc linckia NIES-25 OX=1091006 GN=NIES25_44150 PE=4 SV=1
MM1 pKa = 7.57ANINGTKK8 pKa = 10.23NDD10 pKa = 3.64DD11 pKa = 3.88TLNGSNLPDD20 pKa = 4.43TINGLLGNDD29 pKa = 3.98TITAKK34 pKa = 10.49EE35 pKa = 4.22GNDD38 pKa = 3.64TLSGGSGKK46 pKa = 8.81DD47 pKa = 2.98TFIYY51 pKa = 10.62KK52 pKa = 10.56LGDD55 pKa = 3.41GTDD58 pKa = 4.71FISDD62 pKa = 3.84FGGIGKK68 pKa = 7.92GTNPTAAVIAEE79 pKa = 4.11VDD81 pKa = 3.47TLIFQSYY88 pKa = 9.97GLTARR93 pKa = 11.84NLLLTQNGTNLEE105 pKa = 4.17ITFEE109 pKa = 4.15RR110 pKa = 11.84VDD112 pKa = 3.57DD113 pKa = 4.64AKK115 pKa = 11.4VILQNFALEE124 pKa = 4.15NLDD127 pKa = 3.95NLSKK131 pKa = 10.27STTATVDD138 pKa = 2.93LGNIVFYY145 pKa = 10.27EE146 pKa = 4.21QTSTTDD152 pKa = 2.95SFDD155 pKa = 3.78VFDD158 pKa = 5.19ANSNQSTIFNKK169 pKa = 8.77NTVTFFNDD177 pKa = 2.75LNNNVKK183 pKa = 10.57GFDD186 pKa = 3.67NSDD189 pKa = 3.5DD190 pKa = 4.09VINGQGGNDD199 pKa = 4.06KK200 pKa = 10.69IRR202 pKa = 11.84GLSGNDD208 pKa = 3.55LLRR211 pKa = 11.84GGTGNDD217 pKa = 3.47TLIGGAGNDD226 pKa = 3.92TLVGGAGNNLLNGDD240 pKa = 4.19IDD242 pKa = 5.59DD243 pKa = 5.35DD244 pKa = 4.2LLSIYY249 pKa = 10.16SYY251 pKa = 11.6YY252 pKa = 10.63DD253 pKa = 2.8IYY255 pKa = 11.21DD256 pKa = 3.58YY257 pKa = 10.99PPYY260 pKa = 10.54KK261 pKa = 10.59NNPRR265 pKa = 11.84SDD267 pKa = 3.65GNNTLNGGAGNDD279 pKa = 3.5TLIASRR285 pKa = 11.84STGNNLLDD293 pKa = 4.53GGDD296 pKa = 4.29GNDD299 pKa = 3.58SLDD302 pKa = 3.1ISGYY306 pKa = 10.29DD307 pKa = 3.39EE308 pKa = 4.65YY309 pKa = 11.64VLHH312 pKa = 7.04SDD314 pKa = 4.56YY315 pKa = 11.47SALGNNTLKK324 pKa = 11.02GGAGFDD330 pKa = 3.67NLNANYY336 pKa = 10.57SSGDD340 pKa = 3.45NLLSGGADD348 pKa = 3.42NDD350 pKa = 3.69NLSASGSKK358 pKa = 10.5GNNLLSGGDD367 pKa = 3.52GNDD370 pKa = 3.47YY371 pKa = 11.08VSVSGQYY378 pKa = 9.73QYY380 pKa = 11.75EE381 pKa = 3.89FDD383 pKa = 3.59SYY385 pKa = 11.97GRR387 pKa = 11.84FPTFNFDD394 pKa = 4.58SRR396 pKa = 11.84SLGNNTLNGGNGNDD410 pKa = 3.62TLSASGSKK418 pKa = 10.48GNNLLSGGNGNDD430 pKa = 3.48YY431 pKa = 11.44LNISGSYY438 pKa = 9.51SYY440 pKa = 11.35YY441 pKa = 9.82SNEE444 pKa = 3.99YY445 pKa = 9.19YY446 pKa = 10.51DD447 pKa = 5.65EE448 pKa = 4.53YY449 pKa = 11.3DD450 pKa = 2.91QTYY453 pKa = 9.96YY454 pKa = 11.32YY455 pKa = 10.31SASEE459 pKa = 3.87NDD461 pKa = 3.54SRR463 pKa = 11.84SLGNNTLKK471 pKa = 10.96GGAGDD476 pKa = 3.82DD477 pKa = 3.74TLNAGGSTGNNLLSGGDD494 pKa = 3.58GNDD497 pKa = 3.55YY498 pKa = 10.81ISALNASGNNIIKK511 pKa = 10.36GDD513 pKa = 3.94YY514 pKa = 11.34GNDD517 pKa = 3.32TLNGGFGNDD526 pKa = 3.23TLYY529 pKa = 11.09GGSGIDD535 pKa = 3.17TFGFKK540 pKa = 10.44SFNEE544 pKa = 4.27GLDD547 pKa = 3.41TIYY550 pKa = 11.13DD551 pKa = 3.95FNATNEE557 pKa = 4.25VISIAFGYY565 pKa = 10.73DD566 pKa = 3.52FGAKK570 pKa = 9.44GVLSSTQFTIGTSATTNAQRR590 pKa = 11.84LIYY593 pKa = 10.59NSTTGALFFDD603 pKa = 4.56EE604 pKa = 5.68DD605 pKa = 4.38GSGSTFSQLQFAQLSAGLLLTNNNFVIII633 pKa = 4.2
MM1 pKa = 7.57ANINGTKK8 pKa = 10.23NDD10 pKa = 3.64DD11 pKa = 3.88TLNGSNLPDD20 pKa = 4.43TINGLLGNDD29 pKa = 3.98TITAKK34 pKa = 10.49EE35 pKa = 4.22GNDD38 pKa = 3.64TLSGGSGKK46 pKa = 8.81DD47 pKa = 2.98TFIYY51 pKa = 10.62KK52 pKa = 10.56LGDD55 pKa = 3.41GTDD58 pKa = 4.71FISDD62 pKa = 3.84FGGIGKK68 pKa = 7.92GTNPTAAVIAEE79 pKa = 4.11VDD81 pKa = 3.47TLIFQSYY88 pKa = 9.97GLTARR93 pKa = 11.84NLLLTQNGTNLEE105 pKa = 4.17ITFEE109 pKa = 4.15RR110 pKa = 11.84VDD112 pKa = 3.57DD113 pKa = 4.64AKK115 pKa = 11.4VILQNFALEE124 pKa = 4.15NLDD127 pKa = 3.95NLSKK131 pKa = 10.27STTATVDD138 pKa = 2.93LGNIVFYY145 pKa = 10.27EE146 pKa = 4.21QTSTTDD152 pKa = 2.95SFDD155 pKa = 3.78VFDD158 pKa = 5.19ANSNQSTIFNKK169 pKa = 8.77NTVTFFNDD177 pKa = 2.75LNNNVKK183 pKa = 10.57GFDD186 pKa = 3.67NSDD189 pKa = 3.5DD190 pKa = 4.09VINGQGGNDD199 pKa = 4.06KK200 pKa = 10.69IRR202 pKa = 11.84GLSGNDD208 pKa = 3.55LLRR211 pKa = 11.84GGTGNDD217 pKa = 3.47TLIGGAGNDD226 pKa = 3.92TLVGGAGNNLLNGDD240 pKa = 4.19IDD242 pKa = 5.59DD243 pKa = 5.35DD244 pKa = 4.2LLSIYY249 pKa = 10.16SYY251 pKa = 11.6YY252 pKa = 10.63DD253 pKa = 2.8IYY255 pKa = 11.21DD256 pKa = 3.58YY257 pKa = 10.99PPYY260 pKa = 10.54KK261 pKa = 10.59NNPRR265 pKa = 11.84SDD267 pKa = 3.65GNNTLNGGAGNDD279 pKa = 3.5TLIASRR285 pKa = 11.84STGNNLLDD293 pKa = 4.53GGDD296 pKa = 4.29GNDD299 pKa = 3.58SLDD302 pKa = 3.1ISGYY306 pKa = 10.29DD307 pKa = 3.39EE308 pKa = 4.65YY309 pKa = 11.64VLHH312 pKa = 7.04SDD314 pKa = 4.56YY315 pKa = 11.47SALGNNTLKK324 pKa = 11.02GGAGFDD330 pKa = 3.67NLNANYY336 pKa = 10.57SSGDD340 pKa = 3.45NLLSGGADD348 pKa = 3.42NDD350 pKa = 3.69NLSASGSKK358 pKa = 10.5GNNLLSGGDD367 pKa = 3.52GNDD370 pKa = 3.47YY371 pKa = 11.08VSVSGQYY378 pKa = 9.73QYY380 pKa = 11.75EE381 pKa = 3.89FDD383 pKa = 3.59SYY385 pKa = 11.97GRR387 pKa = 11.84FPTFNFDD394 pKa = 4.58SRR396 pKa = 11.84SLGNNTLNGGNGNDD410 pKa = 3.62TLSASGSKK418 pKa = 10.48GNNLLSGGNGNDD430 pKa = 3.48YY431 pKa = 11.44LNISGSYY438 pKa = 9.51SYY440 pKa = 11.35YY441 pKa = 9.82SNEE444 pKa = 3.99YY445 pKa = 9.19YY446 pKa = 10.51DD447 pKa = 5.65EE448 pKa = 4.53YY449 pKa = 11.3DD450 pKa = 2.91QTYY453 pKa = 9.96YY454 pKa = 11.32YY455 pKa = 10.31SASEE459 pKa = 3.87NDD461 pKa = 3.54SRR463 pKa = 11.84SLGNNTLKK471 pKa = 10.96GGAGDD476 pKa = 3.82DD477 pKa = 3.74TLNAGGSTGNNLLSGGDD494 pKa = 3.58GNDD497 pKa = 3.55YY498 pKa = 10.81ISALNASGNNIIKK511 pKa = 10.36GDD513 pKa = 3.94YY514 pKa = 11.34GNDD517 pKa = 3.32TLNGGFGNDD526 pKa = 3.23TLYY529 pKa = 11.09GGSGIDD535 pKa = 3.17TFGFKK540 pKa = 10.44SFNEE544 pKa = 4.27GLDD547 pKa = 3.41TIYY550 pKa = 11.13DD551 pKa = 3.95FNATNEE557 pKa = 4.25VISIAFGYY565 pKa = 10.73DD566 pKa = 3.52FGAKK570 pKa = 9.44GVLSSTQFTIGTSATTNAQRR590 pKa = 11.84LIYY593 pKa = 10.59NSTTGALFFDD603 pKa = 4.56EE604 pKa = 5.68DD605 pKa = 4.38GSGSTFSQLQFAQLSAGLLLTNNNFVIII633 pKa = 4.2
Molecular weight: 66.46 kDa
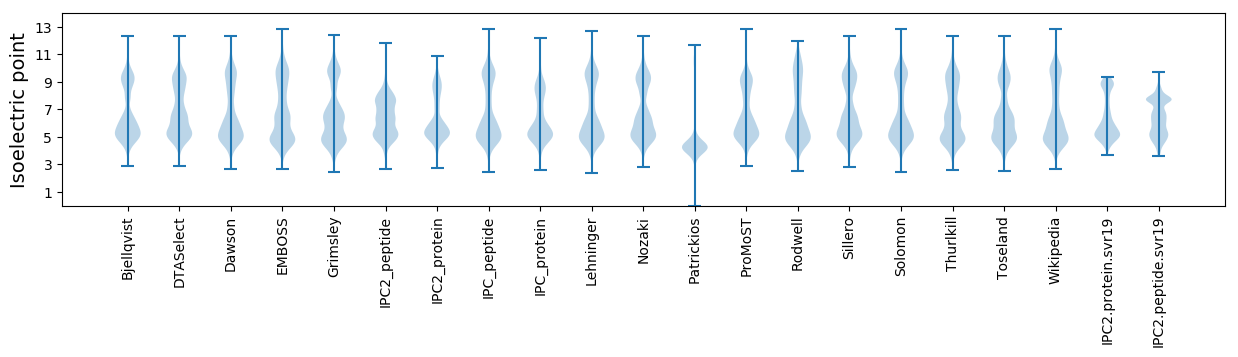
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z4LF21|A0A1Z4LF21_NOSLI 4Fe-4S ferredoxin iron-sulfur binding domain-containing protein OS=Nostoc linckia NIES-25 OX=1091006 GN=NIES25_63120 PE=4 SV=1
MM1 pKa = 7.31KK2 pKa = 10.03PNYY5 pKa = 9.07RR6 pKa = 11.84RR7 pKa = 11.84CISCRR12 pKa = 11.84KK13 pKa = 9.67LGLKK17 pKa = 10.18EE18 pKa = 3.92EE19 pKa = 4.62FWRR22 pKa = 11.84IVRR25 pKa = 11.84VFPSGKK31 pKa = 8.66VQLDD35 pKa = 3.07RR36 pKa = 11.84GMGRR40 pKa = 11.84SAYY43 pKa = 9.3ICPQVSCLQAAQKK56 pKa = 10.5KK57 pKa = 8.83NRR59 pKa = 11.84LGRR62 pKa = 11.84SLHH65 pKa = 6.06ASVPEE70 pKa = 3.79TLYY73 pKa = 10.86QSLSQRR79 pKa = 11.84LASNNTQNQII89 pKa = 3.34
MM1 pKa = 7.31KK2 pKa = 10.03PNYY5 pKa = 9.07RR6 pKa = 11.84RR7 pKa = 11.84CISCRR12 pKa = 11.84KK13 pKa = 9.67LGLKK17 pKa = 10.18EE18 pKa = 3.92EE19 pKa = 4.62FWRR22 pKa = 11.84IVRR25 pKa = 11.84VFPSGKK31 pKa = 8.66VQLDD35 pKa = 3.07RR36 pKa = 11.84GMGRR40 pKa = 11.84SAYY43 pKa = 9.3ICPQVSCLQAAQKK56 pKa = 10.5KK57 pKa = 8.83NRR59 pKa = 11.84LGRR62 pKa = 11.84SLHH65 pKa = 6.06ASVPEE70 pKa = 3.79TLYY73 pKa = 10.86QSLSQRR79 pKa = 11.84LASNNTQNQII89 pKa = 3.34
Molecular weight: 10.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2248921 |
29 |
7234 |
326.8 |
36.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.05 ± 0.03 | 0.942 ± 0.011 |
4.88 ± 0.024 | 6.299 ± 0.034 |
4.032 ± 0.019 | 6.685 ± 0.051 |
1.73 ± 0.014 | 6.957 ± 0.025 |
4.924 ± 0.025 | 10.958 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.694 ± 0.013 | 4.728 ± 0.036 |
4.555 ± 0.025 | 5.379 ± 0.032 |
4.982 ± 0.026 | 6.5 ± 0.026 |
5.726 ± 0.033 | 6.529 ± 0.023 |
1.375 ± 0.014 | 3.077 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
