
Cronartium ribicola mitovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.85
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
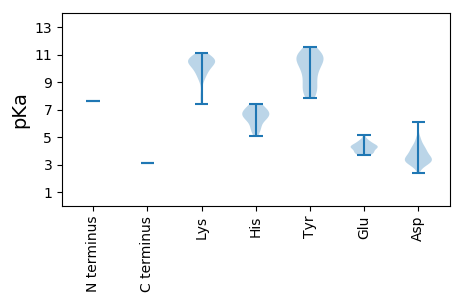
Protein with the lowest isoelectric point:
>tr|A0A191KCP6|A0A191KCP6_9VIRU RNA-dependent RNA polymerase OS=Cronartium ribicola mitovirus 1 OX=1816484 GN=RdRp PE=4 SV=1
MM1 pKa = 7.61RR2 pKa = 11.84MKK4 pKa = 10.65SRR6 pKa = 11.84TWIQASSLLVKK17 pKa = 10.67ISLPKK22 pKa = 10.3DD23 pKa = 3.41LQSCVHH29 pKa = 6.54ILALLSVYY37 pKa = 9.91WEE39 pKa = 4.35IDD41 pKa = 3.3SSDD44 pKa = 3.96WIPYY48 pKa = 8.2IDD50 pKa = 3.32RR51 pKa = 11.84VVKK54 pKa = 10.45IRR56 pKa = 11.84RR57 pKa = 11.84LSGTPYY63 pKa = 7.86TVKK66 pKa = 9.79WNKK69 pKa = 9.53SNRR72 pKa = 11.84LAFTRR77 pKa = 11.84WMCGHH82 pKa = 7.41PLKK85 pKa = 10.93EE86 pKa = 4.28SDD88 pKa = 4.28VPLHH92 pKa = 6.88KK93 pKa = 10.86DD94 pKa = 3.37GFPRR98 pKa = 11.84NLSSMKK104 pKa = 10.17KK105 pKa = 7.39YY106 pKa = 8.89TSSARR111 pKa = 11.84GIAFLLTLLIWSRR124 pKa = 11.84GLHH127 pKa = 5.05QWSKK131 pKa = 10.62PDD133 pKa = 3.12ISAVLSPYY141 pKa = 8.12TGKK144 pKa = 8.06MTEE147 pKa = 4.64DD148 pKa = 2.88KK149 pKa = 10.19WSRR152 pKa = 11.84SPVQFLADD160 pKa = 3.68RR161 pKa = 11.84VRR163 pKa = 11.84LSLGKK168 pKa = 9.28PLEE171 pKa = 4.42PWSRR175 pKa = 11.84WHH177 pKa = 5.67LTTRR181 pKa = 11.84NGPNGHH187 pKa = 7.15ALTGILEE194 pKa = 4.3DD195 pKa = 4.29LKK197 pKa = 11.11AITGSCIPHH206 pKa = 6.78LTTLGGKK213 pKa = 10.11KK214 pKa = 9.25IVTYY218 pKa = 10.64LSHH221 pKa = 7.29FKK223 pKa = 11.0VLTSKK228 pKa = 10.7EE229 pKa = 3.72GALNYY234 pKa = 9.91IKK236 pKa = 10.52EE237 pKa = 4.3RR238 pKa = 11.84LSLRR242 pKa = 11.84YY243 pKa = 9.5QKK245 pKa = 10.54EE246 pKa = 4.36SPSRR250 pKa = 11.84LAKK253 pKa = 10.68LIFIKK258 pKa = 10.73DD259 pKa = 3.47KK260 pKa = 9.06EE261 pKa = 4.3LKK263 pKa = 9.16MRR265 pKa = 11.84SIGIGNYY272 pKa = 8.37WYY274 pKa = 10.44QAALKK279 pKa = 9.63NLHH282 pKa = 6.78DD283 pKa = 3.96QLIDD287 pKa = 3.05ILRR290 pKa = 11.84RR291 pKa = 11.84IPEE294 pKa = 4.64DD295 pKa = 3.21ATFDD299 pKa = 3.32QGKK302 pKa = 9.74APRR305 pKa = 11.84VLRR308 pKa = 11.84KK309 pKa = 9.79PEE311 pKa = 3.82GHH313 pKa = 6.48SYY315 pKa = 10.48WSMDD319 pKa = 3.0LTAATDD325 pKa = 4.88RR326 pKa = 11.84FPLWYY331 pKa = 8.23QKK333 pKa = 11.05KK334 pKa = 10.41VIGEE338 pKa = 4.15LFSAEE343 pKa = 4.17VAEE346 pKa = 4.23SWSYY350 pKa = 11.23IITTKK355 pKa = 10.45FSSRR359 pKa = 11.84EE360 pKa = 3.81FGEE363 pKa = 3.85VKK365 pKa = 10.58YY366 pKa = 10.88AVGQPIGFYY375 pKa = 10.66SSWAAFAVSHH385 pKa = 6.63HH386 pKa = 6.22IVVHH390 pKa = 5.79AAAIKK395 pKa = 10.62AGIRR399 pKa = 11.84DD400 pKa = 3.84LKK402 pKa = 10.69GLYY405 pKa = 9.82VLLGDD410 pKa = 6.08DD411 pKa = 4.89IVLCHH416 pKa = 7.4DD417 pKa = 4.18DD418 pKa = 3.5LARR421 pKa = 11.84EE422 pKa = 3.97YY423 pKa = 10.84KK424 pKa = 10.34IIIGLLGVEE433 pKa = 4.23LSISKK438 pKa = 7.58THH440 pKa = 6.37QSDD443 pKa = 3.12HH444 pKa = 6.23TYY446 pKa = 10.85EE447 pKa = 4.14FAKK450 pKa = 10.1RR451 pKa = 11.84WYY453 pKa = 8.35SDD455 pKa = 2.38KK456 pKa = 10.55WGEE459 pKa = 3.78ISPFPIHH466 pKa = 6.74SVSPKK471 pKa = 10.62AKK473 pKa = 8.2YY474 pKa = 8.85TDD476 pKa = 3.61IIAIINSAAEE486 pKa = 4.44KK487 pKa = 9.81GWSPSSQHH495 pKa = 5.45YY496 pKa = 11.06LEE498 pKa = 5.15FAAASNCLLHH508 pKa = 6.96KK509 pKa = 10.4NHH511 pKa = 6.74GDD513 pKa = 3.31EE514 pKa = 4.77FYY516 pKa = 11.14SRR518 pKa = 11.84SLRR521 pKa = 11.84FLNLYY526 pKa = 8.86RR527 pKa = 11.84LCFIASTGQGSWSEE541 pKa = 4.28VIRR544 pKa = 11.84NLDD547 pKa = 3.23VARR550 pKa = 11.84RR551 pKa = 11.84TISVAGSAWYY561 pKa = 9.11HH562 pKa = 5.55PKK564 pKa = 10.66YY565 pKa = 11.02GEE567 pKa = 4.77DD568 pKa = 3.85LYY570 pKa = 11.51QLTLLVIFLNKK581 pKa = 10.2GVGKK585 pKa = 10.01LATATKK591 pKa = 9.4VASRR595 pKa = 11.84TILNLIPRR603 pKa = 11.84TAEE606 pKa = 3.73PNHH609 pKa = 6.22PCKK612 pKa = 10.68GLWSVLINLPQSHH625 pKa = 6.44IVSDD629 pKa = 3.76IVDD632 pKa = 3.97SFSKK636 pKa = 10.85DD637 pKa = 3.08SFLSLIQQYY646 pKa = 10.27IGQDD650 pKa = 3.32NKK652 pKa = 10.91KK653 pKa = 9.71LSSLITCDD661 pKa = 3.59LNTVLTTRR669 pKa = 11.84NQQLSWNVLHH679 pKa = 6.96KK680 pKa = 10.37VVQAIVVTISRR691 pKa = 11.84VSEE694 pKa = 3.87QGCTFRR700 pKa = 11.84GPEE703 pKa = 3.93QLPLEE708 pKa = 4.56VVRR711 pKa = 11.84AAFKK715 pKa = 10.25PISNRR720 pKa = 11.84SLAVGTRR727 pKa = 11.84YY728 pKa = 9.96VSSDD732 pKa = 3.31NPLYY736 pKa = 10.79SYY738 pKa = 9.42WVEE741 pKa = 3.99MKK743 pKa = 10.45KK744 pKa = 10.65LGVIGQTQMWVTAKK758 pKa = 10.01TKK760 pKa = 10.37GLQPLQQDD768 pKa = 4.75TILDD772 pKa = 4.24DD773 pKa = 4.34PDD775 pKa = 4.2FSIFSRR781 pKa = 11.84IDD783 pKa = 3.33YY784 pKa = 10.42LDD786 pKa = 3.21KK787 pKa = 10.41TGKK790 pKa = 9.75VSSYY794 pKa = 11.08LYY796 pKa = 10.72GEE798 pKa = 4.5RR799 pKa = 11.84GG800 pKa = 3.12
MM1 pKa = 7.61RR2 pKa = 11.84MKK4 pKa = 10.65SRR6 pKa = 11.84TWIQASSLLVKK17 pKa = 10.67ISLPKK22 pKa = 10.3DD23 pKa = 3.41LQSCVHH29 pKa = 6.54ILALLSVYY37 pKa = 9.91WEE39 pKa = 4.35IDD41 pKa = 3.3SSDD44 pKa = 3.96WIPYY48 pKa = 8.2IDD50 pKa = 3.32RR51 pKa = 11.84VVKK54 pKa = 10.45IRR56 pKa = 11.84RR57 pKa = 11.84LSGTPYY63 pKa = 7.86TVKK66 pKa = 9.79WNKK69 pKa = 9.53SNRR72 pKa = 11.84LAFTRR77 pKa = 11.84WMCGHH82 pKa = 7.41PLKK85 pKa = 10.93EE86 pKa = 4.28SDD88 pKa = 4.28VPLHH92 pKa = 6.88KK93 pKa = 10.86DD94 pKa = 3.37GFPRR98 pKa = 11.84NLSSMKK104 pKa = 10.17KK105 pKa = 7.39YY106 pKa = 8.89TSSARR111 pKa = 11.84GIAFLLTLLIWSRR124 pKa = 11.84GLHH127 pKa = 5.05QWSKK131 pKa = 10.62PDD133 pKa = 3.12ISAVLSPYY141 pKa = 8.12TGKK144 pKa = 8.06MTEE147 pKa = 4.64DD148 pKa = 2.88KK149 pKa = 10.19WSRR152 pKa = 11.84SPVQFLADD160 pKa = 3.68RR161 pKa = 11.84VRR163 pKa = 11.84LSLGKK168 pKa = 9.28PLEE171 pKa = 4.42PWSRR175 pKa = 11.84WHH177 pKa = 5.67LTTRR181 pKa = 11.84NGPNGHH187 pKa = 7.15ALTGILEE194 pKa = 4.3DD195 pKa = 4.29LKK197 pKa = 11.11AITGSCIPHH206 pKa = 6.78LTTLGGKK213 pKa = 10.11KK214 pKa = 9.25IVTYY218 pKa = 10.64LSHH221 pKa = 7.29FKK223 pKa = 11.0VLTSKK228 pKa = 10.7EE229 pKa = 3.72GALNYY234 pKa = 9.91IKK236 pKa = 10.52EE237 pKa = 4.3RR238 pKa = 11.84LSLRR242 pKa = 11.84YY243 pKa = 9.5QKK245 pKa = 10.54EE246 pKa = 4.36SPSRR250 pKa = 11.84LAKK253 pKa = 10.68LIFIKK258 pKa = 10.73DD259 pKa = 3.47KK260 pKa = 9.06EE261 pKa = 4.3LKK263 pKa = 9.16MRR265 pKa = 11.84SIGIGNYY272 pKa = 8.37WYY274 pKa = 10.44QAALKK279 pKa = 9.63NLHH282 pKa = 6.78DD283 pKa = 3.96QLIDD287 pKa = 3.05ILRR290 pKa = 11.84RR291 pKa = 11.84IPEE294 pKa = 4.64DD295 pKa = 3.21ATFDD299 pKa = 3.32QGKK302 pKa = 9.74APRR305 pKa = 11.84VLRR308 pKa = 11.84KK309 pKa = 9.79PEE311 pKa = 3.82GHH313 pKa = 6.48SYY315 pKa = 10.48WSMDD319 pKa = 3.0LTAATDD325 pKa = 4.88RR326 pKa = 11.84FPLWYY331 pKa = 8.23QKK333 pKa = 11.05KK334 pKa = 10.41VIGEE338 pKa = 4.15LFSAEE343 pKa = 4.17VAEE346 pKa = 4.23SWSYY350 pKa = 11.23IITTKK355 pKa = 10.45FSSRR359 pKa = 11.84EE360 pKa = 3.81FGEE363 pKa = 3.85VKK365 pKa = 10.58YY366 pKa = 10.88AVGQPIGFYY375 pKa = 10.66SSWAAFAVSHH385 pKa = 6.63HH386 pKa = 6.22IVVHH390 pKa = 5.79AAAIKK395 pKa = 10.62AGIRR399 pKa = 11.84DD400 pKa = 3.84LKK402 pKa = 10.69GLYY405 pKa = 9.82VLLGDD410 pKa = 6.08DD411 pKa = 4.89IVLCHH416 pKa = 7.4DD417 pKa = 4.18DD418 pKa = 3.5LARR421 pKa = 11.84EE422 pKa = 3.97YY423 pKa = 10.84KK424 pKa = 10.34IIIGLLGVEE433 pKa = 4.23LSISKK438 pKa = 7.58THH440 pKa = 6.37QSDD443 pKa = 3.12HH444 pKa = 6.23TYY446 pKa = 10.85EE447 pKa = 4.14FAKK450 pKa = 10.1RR451 pKa = 11.84WYY453 pKa = 8.35SDD455 pKa = 2.38KK456 pKa = 10.55WGEE459 pKa = 3.78ISPFPIHH466 pKa = 6.74SVSPKK471 pKa = 10.62AKK473 pKa = 8.2YY474 pKa = 8.85TDD476 pKa = 3.61IIAIINSAAEE486 pKa = 4.44KK487 pKa = 9.81GWSPSSQHH495 pKa = 5.45YY496 pKa = 11.06LEE498 pKa = 5.15FAAASNCLLHH508 pKa = 6.96KK509 pKa = 10.4NHH511 pKa = 6.74GDD513 pKa = 3.31EE514 pKa = 4.77FYY516 pKa = 11.14SRR518 pKa = 11.84SLRR521 pKa = 11.84FLNLYY526 pKa = 8.86RR527 pKa = 11.84LCFIASTGQGSWSEE541 pKa = 4.28VIRR544 pKa = 11.84NLDD547 pKa = 3.23VARR550 pKa = 11.84RR551 pKa = 11.84TISVAGSAWYY561 pKa = 9.11HH562 pKa = 5.55PKK564 pKa = 10.66YY565 pKa = 11.02GEE567 pKa = 4.77DD568 pKa = 3.85LYY570 pKa = 11.51QLTLLVIFLNKK581 pKa = 10.2GVGKK585 pKa = 10.01LATATKK591 pKa = 9.4VASRR595 pKa = 11.84TILNLIPRR603 pKa = 11.84TAEE606 pKa = 3.73PNHH609 pKa = 6.22PCKK612 pKa = 10.68GLWSVLINLPQSHH625 pKa = 6.44IVSDD629 pKa = 3.76IVDD632 pKa = 3.97SFSKK636 pKa = 10.85DD637 pKa = 3.08SFLSLIQQYY646 pKa = 10.27IGQDD650 pKa = 3.32NKK652 pKa = 10.91KK653 pKa = 9.71LSSLITCDD661 pKa = 3.59LNTVLTTRR669 pKa = 11.84NQQLSWNVLHH679 pKa = 6.96KK680 pKa = 10.37VVQAIVVTISRR691 pKa = 11.84VSEE694 pKa = 3.87QGCTFRR700 pKa = 11.84GPEE703 pKa = 3.93QLPLEE708 pKa = 4.56VVRR711 pKa = 11.84AAFKK715 pKa = 10.25PISNRR720 pKa = 11.84SLAVGTRR727 pKa = 11.84YY728 pKa = 9.96VSSDD732 pKa = 3.31NPLYY736 pKa = 10.79SYY738 pKa = 9.42WVEE741 pKa = 3.99MKK743 pKa = 10.45KK744 pKa = 10.65LGVIGQTQMWVTAKK758 pKa = 10.01TKK760 pKa = 10.37GLQPLQQDD768 pKa = 4.75TILDD772 pKa = 4.24DD773 pKa = 4.34PDD775 pKa = 4.2FSIFSRR781 pKa = 11.84IDD783 pKa = 3.33YY784 pKa = 10.42LDD786 pKa = 3.21KK787 pKa = 10.41TGKK790 pKa = 9.75VSSYY794 pKa = 11.08LYY796 pKa = 10.72GEE798 pKa = 4.5RR799 pKa = 11.84GG800 pKa = 3.12
Molecular weight: 90.71 kDa
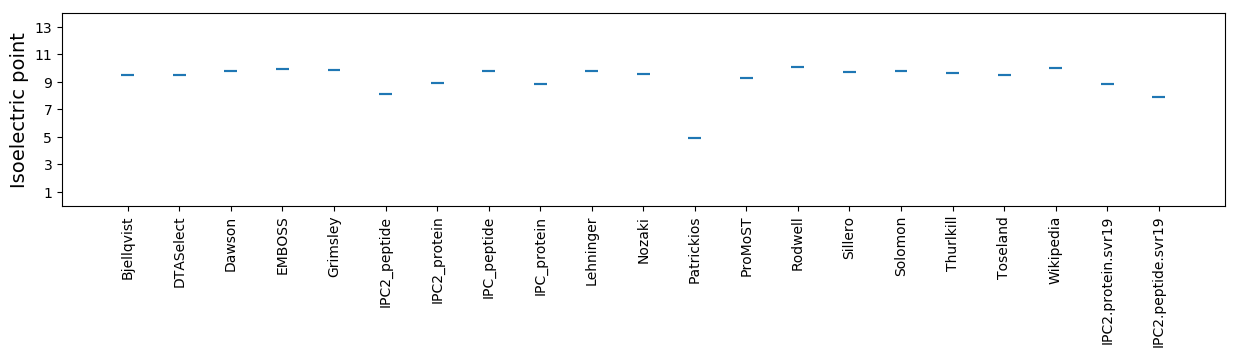
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A191KCP6|A0A191KCP6_9VIRU RNA-dependent RNA polymerase OS=Cronartium ribicola mitovirus 1 OX=1816484 GN=RdRp PE=4 SV=1
MM1 pKa = 7.61RR2 pKa = 11.84MKK4 pKa = 10.65SRR6 pKa = 11.84TWIQASSLLVKK17 pKa = 10.67ISLPKK22 pKa = 10.3DD23 pKa = 3.41LQSCVHH29 pKa = 6.54ILALLSVYY37 pKa = 9.91WEE39 pKa = 4.35IDD41 pKa = 3.3SSDD44 pKa = 3.96WIPYY48 pKa = 8.2IDD50 pKa = 3.32RR51 pKa = 11.84VVKK54 pKa = 10.45IRR56 pKa = 11.84RR57 pKa = 11.84LSGTPYY63 pKa = 7.86TVKK66 pKa = 9.79WNKK69 pKa = 9.53SNRR72 pKa = 11.84LAFTRR77 pKa = 11.84WMCGHH82 pKa = 7.41PLKK85 pKa = 10.93EE86 pKa = 4.28SDD88 pKa = 4.28VPLHH92 pKa = 6.88KK93 pKa = 10.86DD94 pKa = 3.37GFPRR98 pKa = 11.84NLSSMKK104 pKa = 10.17KK105 pKa = 7.39YY106 pKa = 8.89TSSARR111 pKa = 11.84GIAFLLTLLIWSRR124 pKa = 11.84GLHH127 pKa = 5.05QWSKK131 pKa = 10.62PDD133 pKa = 3.12ISAVLSPYY141 pKa = 8.12TGKK144 pKa = 8.06MTEE147 pKa = 4.64DD148 pKa = 2.88KK149 pKa = 10.19WSRR152 pKa = 11.84SPVQFLADD160 pKa = 3.68RR161 pKa = 11.84VRR163 pKa = 11.84LSLGKK168 pKa = 9.28PLEE171 pKa = 4.42PWSRR175 pKa = 11.84WHH177 pKa = 5.67LTTRR181 pKa = 11.84NGPNGHH187 pKa = 7.15ALTGILEE194 pKa = 4.3DD195 pKa = 4.29LKK197 pKa = 11.11AITGSCIPHH206 pKa = 6.78LTTLGGKK213 pKa = 10.11KK214 pKa = 9.25IVTYY218 pKa = 10.64LSHH221 pKa = 7.29FKK223 pKa = 11.0VLTSKK228 pKa = 10.7EE229 pKa = 3.72GALNYY234 pKa = 9.91IKK236 pKa = 10.52EE237 pKa = 4.3RR238 pKa = 11.84LSLRR242 pKa = 11.84YY243 pKa = 9.5QKK245 pKa = 10.54EE246 pKa = 4.36SPSRR250 pKa = 11.84LAKK253 pKa = 10.68LIFIKK258 pKa = 10.73DD259 pKa = 3.47KK260 pKa = 9.06EE261 pKa = 4.3LKK263 pKa = 9.16MRR265 pKa = 11.84SIGIGNYY272 pKa = 8.37WYY274 pKa = 10.44QAALKK279 pKa = 9.63NLHH282 pKa = 6.78DD283 pKa = 3.96QLIDD287 pKa = 3.05ILRR290 pKa = 11.84RR291 pKa = 11.84IPEE294 pKa = 4.64DD295 pKa = 3.21ATFDD299 pKa = 3.32QGKK302 pKa = 9.74APRR305 pKa = 11.84VLRR308 pKa = 11.84KK309 pKa = 9.79PEE311 pKa = 3.82GHH313 pKa = 6.48SYY315 pKa = 10.48WSMDD319 pKa = 3.0LTAATDD325 pKa = 4.88RR326 pKa = 11.84FPLWYY331 pKa = 8.23QKK333 pKa = 11.05KK334 pKa = 10.41VIGEE338 pKa = 4.15LFSAEE343 pKa = 4.17VAEE346 pKa = 4.23SWSYY350 pKa = 11.23IITTKK355 pKa = 10.45FSSRR359 pKa = 11.84EE360 pKa = 3.81FGEE363 pKa = 3.85VKK365 pKa = 10.58YY366 pKa = 10.88AVGQPIGFYY375 pKa = 10.66SSWAAFAVSHH385 pKa = 6.63HH386 pKa = 6.22IVVHH390 pKa = 5.79AAAIKK395 pKa = 10.62AGIRR399 pKa = 11.84DD400 pKa = 3.84LKK402 pKa = 10.69GLYY405 pKa = 9.82VLLGDD410 pKa = 6.08DD411 pKa = 4.89IVLCHH416 pKa = 7.4DD417 pKa = 4.18DD418 pKa = 3.5LARR421 pKa = 11.84EE422 pKa = 3.97YY423 pKa = 10.84KK424 pKa = 10.34IIIGLLGVEE433 pKa = 4.23LSISKK438 pKa = 7.58THH440 pKa = 6.37QSDD443 pKa = 3.12HH444 pKa = 6.23TYY446 pKa = 10.85EE447 pKa = 4.14FAKK450 pKa = 10.1RR451 pKa = 11.84WYY453 pKa = 8.35SDD455 pKa = 2.38KK456 pKa = 10.55WGEE459 pKa = 3.78ISPFPIHH466 pKa = 6.74SVSPKK471 pKa = 10.62AKK473 pKa = 8.2YY474 pKa = 8.85TDD476 pKa = 3.61IIAIINSAAEE486 pKa = 4.44KK487 pKa = 9.81GWSPSSQHH495 pKa = 5.45YY496 pKa = 11.06LEE498 pKa = 5.15FAAASNCLLHH508 pKa = 6.96KK509 pKa = 10.4NHH511 pKa = 6.74GDD513 pKa = 3.31EE514 pKa = 4.77FYY516 pKa = 11.14SRR518 pKa = 11.84SLRR521 pKa = 11.84FLNLYY526 pKa = 8.86RR527 pKa = 11.84LCFIASTGQGSWSEE541 pKa = 4.28VIRR544 pKa = 11.84NLDD547 pKa = 3.23VARR550 pKa = 11.84RR551 pKa = 11.84TISVAGSAWYY561 pKa = 9.11HH562 pKa = 5.55PKK564 pKa = 10.66YY565 pKa = 11.02GEE567 pKa = 4.77DD568 pKa = 3.85LYY570 pKa = 11.51QLTLLVIFLNKK581 pKa = 10.2GVGKK585 pKa = 10.01LATATKK591 pKa = 9.4VASRR595 pKa = 11.84TILNLIPRR603 pKa = 11.84TAEE606 pKa = 3.73PNHH609 pKa = 6.22PCKK612 pKa = 10.68GLWSVLINLPQSHH625 pKa = 6.44IVSDD629 pKa = 3.76IVDD632 pKa = 3.97SFSKK636 pKa = 10.85DD637 pKa = 3.08SFLSLIQQYY646 pKa = 10.27IGQDD650 pKa = 3.32NKK652 pKa = 10.91KK653 pKa = 9.71LSSLITCDD661 pKa = 3.59LNTVLTTRR669 pKa = 11.84NQQLSWNVLHH679 pKa = 6.96KK680 pKa = 10.37VVQAIVVTISRR691 pKa = 11.84VSEE694 pKa = 3.87QGCTFRR700 pKa = 11.84GPEE703 pKa = 3.93QLPLEE708 pKa = 4.56VVRR711 pKa = 11.84AAFKK715 pKa = 10.25PISNRR720 pKa = 11.84SLAVGTRR727 pKa = 11.84YY728 pKa = 9.96VSSDD732 pKa = 3.31NPLYY736 pKa = 10.79SYY738 pKa = 9.42WVEE741 pKa = 3.99MKK743 pKa = 10.45KK744 pKa = 10.65LGVIGQTQMWVTAKK758 pKa = 10.01TKK760 pKa = 10.37GLQPLQQDD768 pKa = 4.75TILDD772 pKa = 4.24DD773 pKa = 4.34PDD775 pKa = 4.2FSIFSRR781 pKa = 11.84IDD783 pKa = 3.33YY784 pKa = 10.42LDD786 pKa = 3.21KK787 pKa = 10.41TGKK790 pKa = 9.75VSSYY794 pKa = 11.08LYY796 pKa = 10.72GEE798 pKa = 4.5RR799 pKa = 11.84GG800 pKa = 3.12
MM1 pKa = 7.61RR2 pKa = 11.84MKK4 pKa = 10.65SRR6 pKa = 11.84TWIQASSLLVKK17 pKa = 10.67ISLPKK22 pKa = 10.3DD23 pKa = 3.41LQSCVHH29 pKa = 6.54ILALLSVYY37 pKa = 9.91WEE39 pKa = 4.35IDD41 pKa = 3.3SSDD44 pKa = 3.96WIPYY48 pKa = 8.2IDD50 pKa = 3.32RR51 pKa = 11.84VVKK54 pKa = 10.45IRR56 pKa = 11.84RR57 pKa = 11.84LSGTPYY63 pKa = 7.86TVKK66 pKa = 9.79WNKK69 pKa = 9.53SNRR72 pKa = 11.84LAFTRR77 pKa = 11.84WMCGHH82 pKa = 7.41PLKK85 pKa = 10.93EE86 pKa = 4.28SDD88 pKa = 4.28VPLHH92 pKa = 6.88KK93 pKa = 10.86DD94 pKa = 3.37GFPRR98 pKa = 11.84NLSSMKK104 pKa = 10.17KK105 pKa = 7.39YY106 pKa = 8.89TSSARR111 pKa = 11.84GIAFLLTLLIWSRR124 pKa = 11.84GLHH127 pKa = 5.05QWSKK131 pKa = 10.62PDD133 pKa = 3.12ISAVLSPYY141 pKa = 8.12TGKK144 pKa = 8.06MTEE147 pKa = 4.64DD148 pKa = 2.88KK149 pKa = 10.19WSRR152 pKa = 11.84SPVQFLADD160 pKa = 3.68RR161 pKa = 11.84VRR163 pKa = 11.84LSLGKK168 pKa = 9.28PLEE171 pKa = 4.42PWSRR175 pKa = 11.84WHH177 pKa = 5.67LTTRR181 pKa = 11.84NGPNGHH187 pKa = 7.15ALTGILEE194 pKa = 4.3DD195 pKa = 4.29LKK197 pKa = 11.11AITGSCIPHH206 pKa = 6.78LTTLGGKK213 pKa = 10.11KK214 pKa = 9.25IVTYY218 pKa = 10.64LSHH221 pKa = 7.29FKK223 pKa = 11.0VLTSKK228 pKa = 10.7EE229 pKa = 3.72GALNYY234 pKa = 9.91IKK236 pKa = 10.52EE237 pKa = 4.3RR238 pKa = 11.84LSLRR242 pKa = 11.84YY243 pKa = 9.5QKK245 pKa = 10.54EE246 pKa = 4.36SPSRR250 pKa = 11.84LAKK253 pKa = 10.68LIFIKK258 pKa = 10.73DD259 pKa = 3.47KK260 pKa = 9.06EE261 pKa = 4.3LKK263 pKa = 9.16MRR265 pKa = 11.84SIGIGNYY272 pKa = 8.37WYY274 pKa = 10.44QAALKK279 pKa = 9.63NLHH282 pKa = 6.78DD283 pKa = 3.96QLIDD287 pKa = 3.05ILRR290 pKa = 11.84RR291 pKa = 11.84IPEE294 pKa = 4.64DD295 pKa = 3.21ATFDD299 pKa = 3.32QGKK302 pKa = 9.74APRR305 pKa = 11.84VLRR308 pKa = 11.84KK309 pKa = 9.79PEE311 pKa = 3.82GHH313 pKa = 6.48SYY315 pKa = 10.48WSMDD319 pKa = 3.0LTAATDD325 pKa = 4.88RR326 pKa = 11.84FPLWYY331 pKa = 8.23QKK333 pKa = 11.05KK334 pKa = 10.41VIGEE338 pKa = 4.15LFSAEE343 pKa = 4.17VAEE346 pKa = 4.23SWSYY350 pKa = 11.23IITTKK355 pKa = 10.45FSSRR359 pKa = 11.84EE360 pKa = 3.81FGEE363 pKa = 3.85VKK365 pKa = 10.58YY366 pKa = 10.88AVGQPIGFYY375 pKa = 10.66SSWAAFAVSHH385 pKa = 6.63HH386 pKa = 6.22IVVHH390 pKa = 5.79AAAIKK395 pKa = 10.62AGIRR399 pKa = 11.84DD400 pKa = 3.84LKK402 pKa = 10.69GLYY405 pKa = 9.82VLLGDD410 pKa = 6.08DD411 pKa = 4.89IVLCHH416 pKa = 7.4DD417 pKa = 4.18DD418 pKa = 3.5LARR421 pKa = 11.84EE422 pKa = 3.97YY423 pKa = 10.84KK424 pKa = 10.34IIIGLLGVEE433 pKa = 4.23LSISKK438 pKa = 7.58THH440 pKa = 6.37QSDD443 pKa = 3.12HH444 pKa = 6.23TYY446 pKa = 10.85EE447 pKa = 4.14FAKK450 pKa = 10.1RR451 pKa = 11.84WYY453 pKa = 8.35SDD455 pKa = 2.38KK456 pKa = 10.55WGEE459 pKa = 3.78ISPFPIHH466 pKa = 6.74SVSPKK471 pKa = 10.62AKK473 pKa = 8.2YY474 pKa = 8.85TDD476 pKa = 3.61IIAIINSAAEE486 pKa = 4.44KK487 pKa = 9.81GWSPSSQHH495 pKa = 5.45YY496 pKa = 11.06LEE498 pKa = 5.15FAAASNCLLHH508 pKa = 6.96KK509 pKa = 10.4NHH511 pKa = 6.74GDD513 pKa = 3.31EE514 pKa = 4.77FYY516 pKa = 11.14SRR518 pKa = 11.84SLRR521 pKa = 11.84FLNLYY526 pKa = 8.86RR527 pKa = 11.84LCFIASTGQGSWSEE541 pKa = 4.28VIRR544 pKa = 11.84NLDD547 pKa = 3.23VARR550 pKa = 11.84RR551 pKa = 11.84TISVAGSAWYY561 pKa = 9.11HH562 pKa = 5.55PKK564 pKa = 10.66YY565 pKa = 11.02GEE567 pKa = 4.77DD568 pKa = 3.85LYY570 pKa = 11.51QLTLLVIFLNKK581 pKa = 10.2GVGKK585 pKa = 10.01LATATKK591 pKa = 9.4VASRR595 pKa = 11.84TILNLIPRR603 pKa = 11.84TAEE606 pKa = 3.73PNHH609 pKa = 6.22PCKK612 pKa = 10.68GLWSVLINLPQSHH625 pKa = 6.44IVSDD629 pKa = 3.76IVDD632 pKa = 3.97SFSKK636 pKa = 10.85DD637 pKa = 3.08SFLSLIQQYY646 pKa = 10.27IGQDD650 pKa = 3.32NKK652 pKa = 10.91KK653 pKa = 9.71LSSLITCDD661 pKa = 3.59LNTVLTTRR669 pKa = 11.84NQQLSWNVLHH679 pKa = 6.96KK680 pKa = 10.37VVQAIVVTISRR691 pKa = 11.84VSEE694 pKa = 3.87QGCTFRR700 pKa = 11.84GPEE703 pKa = 3.93QLPLEE708 pKa = 4.56VVRR711 pKa = 11.84AAFKK715 pKa = 10.25PISNRR720 pKa = 11.84SLAVGTRR727 pKa = 11.84YY728 pKa = 9.96VSSDD732 pKa = 3.31NPLYY736 pKa = 10.79SYY738 pKa = 9.42WVEE741 pKa = 3.99MKK743 pKa = 10.45KK744 pKa = 10.65LGVIGQTQMWVTAKK758 pKa = 10.01TKK760 pKa = 10.37GLQPLQQDD768 pKa = 4.75TILDD772 pKa = 4.24DD773 pKa = 4.34PDD775 pKa = 4.2FSIFSRR781 pKa = 11.84IDD783 pKa = 3.33YY784 pKa = 10.42LDD786 pKa = 3.21KK787 pKa = 10.41TGKK790 pKa = 9.75VSSYY794 pKa = 11.08LYY796 pKa = 10.72GEE798 pKa = 4.5RR799 pKa = 11.84GG800 pKa = 3.12
Molecular weight: 90.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
800 |
800 |
800 |
800.0 |
90.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.125 ± 0.0 | 1.125 ± 0.0 |
5.0 ± 0.0 | 3.875 ± 0.0 |
3.25 ± 0.0 | 5.875 ± 0.0 |
3.0 ± 0.0 | 7.375 ± 0.0 |
7.25 ± 0.0 | 11.25 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.125 ± 0.0 | 2.875 ± 0.0 |
4.25 ± 0.0 | 3.5 ± 0.0 |
5.5 ± 0.0 | 10.125 ± 0.0 |
5.375 ± 0.0 | 6.0 ± 0.0 |
3.0 ± 0.0 | 4.125 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
