
Syntrophotalea acetylenivorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfuromonadales; Syntrophotaleaceae; Syntrophotalea
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
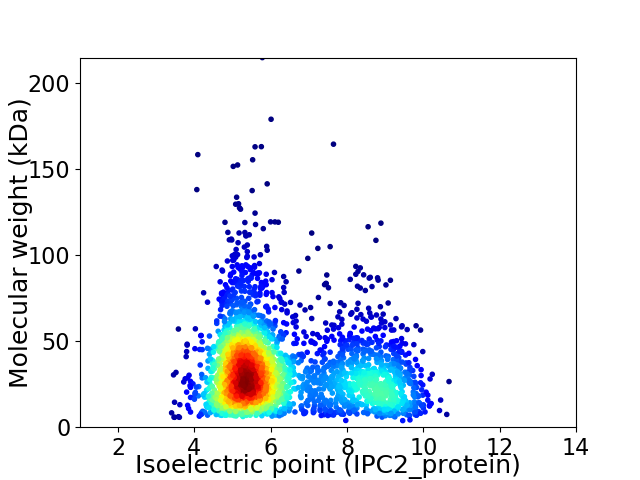
Virtual 2D-PAGE plot for 2762 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3GRU3|A0A1L3GRU3_9DELT Hydroxylamine reductase OS=Syntrophotalea acetylenivorans OX=1842532 GN=hcp PE=3 SV=1
MM1 pKa = 7.93IITGLLIPAVALAEE15 pKa = 4.22LKK17 pKa = 10.0TGVVTVSPMLGGVLMEE33 pKa = 5.1GDD35 pKa = 3.95QPVDD39 pKa = 3.66DD40 pKa = 5.73DD41 pKa = 4.26GVAYY45 pKa = 10.65SLGLGYY51 pKa = 10.68NLSKK55 pKa = 10.93EE56 pKa = 4.33FGLEE60 pKa = 3.85AVLGGANLDD69 pKa = 4.07SDD71 pKa = 4.83DD72 pKa = 5.52SSDD75 pKa = 3.79SDD77 pKa = 3.41VDD79 pKa = 3.96LFTYY83 pKa = 10.61RR84 pKa = 11.84LDD86 pKa = 3.27ALYY89 pKa = 11.01RR90 pKa = 11.84FMPDD94 pKa = 2.83NKK96 pKa = 10.16LVPYY100 pKa = 9.98LAAGVGGYY108 pKa = 10.49DD109 pKa = 4.92LDD111 pKa = 5.19NDD113 pKa = 4.54HH114 pKa = 6.7EE115 pKa = 4.44WMANYY120 pKa = 10.38GGGLLYY126 pKa = 10.33FFAEE130 pKa = 4.65NIALRR135 pKa = 11.84ADD137 pKa = 3.61VRR139 pKa = 11.84HH140 pKa = 6.19LMAFNEE146 pKa = 4.49SNLEE150 pKa = 4.03HH151 pKa = 6.86NLTYY155 pKa = 10.03TAGFLVQFAGVTAPAPKK172 pKa = 10.44APLDD176 pKa = 3.7SDD178 pKa = 3.91GDD180 pKa = 4.3GVTDD184 pKa = 6.62DD185 pKa = 6.29LDD187 pKa = 3.59QCPNTPQGAPVDD199 pKa = 4.11TKK201 pKa = 11.29GCPLDD206 pKa = 3.72SDD208 pKa = 4.0GDD210 pKa = 4.31GVFDD214 pKa = 5.93YY215 pKa = 11.29LDD217 pKa = 3.54QCPDD221 pKa = 3.44TPADD225 pKa = 3.59APVDD229 pKa = 3.86NKK231 pKa = 10.9GCPLDD236 pKa = 3.8SDD238 pKa = 4.0GDD240 pKa = 4.31GVFDD244 pKa = 5.93YY245 pKa = 11.29LDD247 pKa = 3.54QCPDD251 pKa = 3.43TPAGAPVDD259 pKa = 4.21SKK261 pKa = 11.7GCSLDD266 pKa = 3.35SDD268 pKa = 3.96GDD270 pKa = 3.9GVYY273 pKa = 10.23DD274 pKa = 5.54YY275 pKa = 11.55LDD277 pKa = 3.52QCPGTPAGIPVDD289 pKa = 3.62ATGCEE294 pKa = 4.02LKK296 pKa = 10.02LTLRR300 pKa = 11.84INFDD304 pKa = 3.67FNSAVIKK311 pKa = 10.56PEE313 pKa = 3.74FNAEE317 pKa = 3.43VDD319 pKa = 3.39KK320 pKa = 11.31AAEE323 pKa = 3.91FVRR326 pKa = 11.84ANKK329 pKa = 9.58NVPFILLTGHH339 pKa = 6.42TDD341 pKa = 3.21SMGKK345 pKa = 8.82EE346 pKa = 4.22AYY348 pKa = 9.46NQQLSDD354 pKa = 3.31QRR356 pKa = 11.84AAAVRR361 pKa = 11.84QALIDD366 pKa = 3.45QYY368 pKa = 11.87GFDD371 pKa = 4.01GDD373 pKa = 3.93KK374 pKa = 10.83LVARR378 pKa = 11.84GYY380 pKa = 11.52GEE382 pKa = 4.13TQPIADD388 pKa = 3.79NSTEE392 pKa = 3.7EE393 pKa = 3.75GRR395 pKa = 11.84YY396 pKa = 6.55QNRR399 pKa = 11.84RR400 pKa = 11.84VEE402 pKa = 4.24LVCCAVLPEE411 pKa = 4.05
MM1 pKa = 7.93IITGLLIPAVALAEE15 pKa = 4.22LKK17 pKa = 10.0TGVVTVSPMLGGVLMEE33 pKa = 5.1GDD35 pKa = 3.95QPVDD39 pKa = 3.66DD40 pKa = 5.73DD41 pKa = 4.26GVAYY45 pKa = 10.65SLGLGYY51 pKa = 10.68NLSKK55 pKa = 10.93EE56 pKa = 4.33FGLEE60 pKa = 3.85AVLGGANLDD69 pKa = 4.07SDD71 pKa = 4.83DD72 pKa = 5.52SSDD75 pKa = 3.79SDD77 pKa = 3.41VDD79 pKa = 3.96LFTYY83 pKa = 10.61RR84 pKa = 11.84LDD86 pKa = 3.27ALYY89 pKa = 11.01RR90 pKa = 11.84FMPDD94 pKa = 2.83NKK96 pKa = 10.16LVPYY100 pKa = 9.98LAAGVGGYY108 pKa = 10.49DD109 pKa = 4.92LDD111 pKa = 5.19NDD113 pKa = 4.54HH114 pKa = 6.7EE115 pKa = 4.44WMANYY120 pKa = 10.38GGGLLYY126 pKa = 10.33FFAEE130 pKa = 4.65NIALRR135 pKa = 11.84ADD137 pKa = 3.61VRR139 pKa = 11.84HH140 pKa = 6.19LMAFNEE146 pKa = 4.49SNLEE150 pKa = 4.03HH151 pKa = 6.86NLTYY155 pKa = 10.03TAGFLVQFAGVTAPAPKK172 pKa = 10.44APLDD176 pKa = 3.7SDD178 pKa = 3.91GDD180 pKa = 4.3GVTDD184 pKa = 6.62DD185 pKa = 6.29LDD187 pKa = 3.59QCPNTPQGAPVDD199 pKa = 4.11TKK201 pKa = 11.29GCPLDD206 pKa = 3.72SDD208 pKa = 4.0GDD210 pKa = 4.31GVFDD214 pKa = 5.93YY215 pKa = 11.29LDD217 pKa = 3.54QCPDD221 pKa = 3.44TPADD225 pKa = 3.59APVDD229 pKa = 3.86NKK231 pKa = 10.9GCPLDD236 pKa = 3.8SDD238 pKa = 4.0GDD240 pKa = 4.31GVFDD244 pKa = 5.93YY245 pKa = 11.29LDD247 pKa = 3.54QCPDD251 pKa = 3.43TPAGAPVDD259 pKa = 4.21SKK261 pKa = 11.7GCSLDD266 pKa = 3.35SDD268 pKa = 3.96GDD270 pKa = 3.9GVYY273 pKa = 10.23DD274 pKa = 5.54YY275 pKa = 11.55LDD277 pKa = 3.52QCPGTPAGIPVDD289 pKa = 3.62ATGCEE294 pKa = 4.02LKK296 pKa = 10.02LTLRR300 pKa = 11.84INFDD304 pKa = 3.67FNSAVIKK311 pKa = 10.56PEE313 pKa = 3.74FNAEE317 pKa = 3.43VDD319 pKa = 3.39KK320 pKa = 11.31AAEE323 pKa = 3.91FVRR326 pKa = 11.84ANKK329 pKa = 9.58NVPFILLTGHH339 pKa = 6.42TDD341 pKa = 3.21SMGKK345 pKa = 8.82EE346 pKa = 4.22AYY348 pKa = 9.46NQQLSDD354 pKa = 3.31QRR356 pKa = 11.84AAAVRR361 pKa = 11.84QALIDD366 pKa = 3.45QYY368 pKa = 11.87GFDD371 pKa = 4.01GDD373 pKa = 3.93KK374 pKa = 10.83LVARR378 pKa = 11.84GYY380 pKa = 11.52GEE382 pKa = 4.13TQPIADD388 pKa = 3.79NSTEE392 pKa = 3.7EE393 pKa = 3.75GRR395 pKa = 11.84YY396 pKa = 6.55QNRR399 pKa = 11.84RR400 pKa = 11.84VEE402 pKa = 4.24LVCCAVLPEE411 pKa = 4.05
Molecular weight: 43.89 kDa
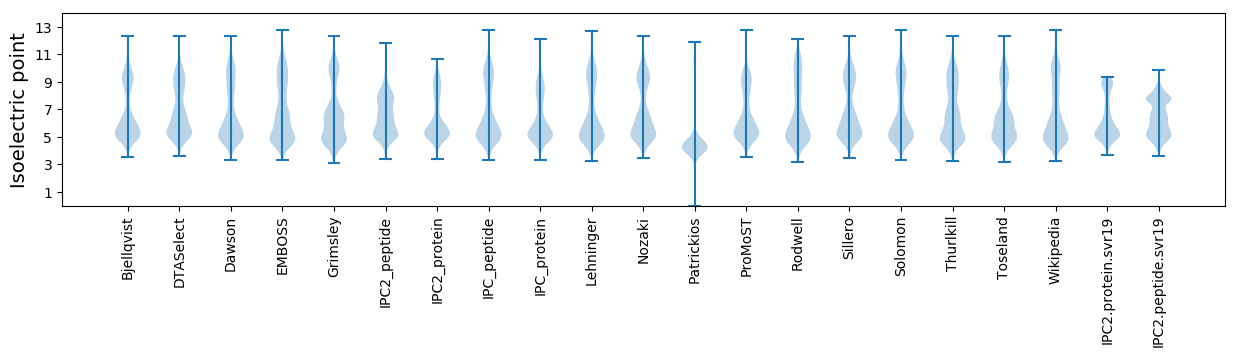
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3GR09|A0A1L3GR09_9DELT Transketolase OS=Syntrophotalea acetylenivorans OX=1842532 GN=A7E78_11150 PE=4 SV=1
MM1 pKa = 7.3FFSLKK6 pKa = 9.98IALSSLRR13 pKa = 11.84AHH15 pKa = 7.57RR16 pKa = 11.84LRR18 pKa = 11.84SILAMVGVFLGALALTGVQHH38 pKa = 6.5VSRR41 pKa = 11.84SMLLKK46 pKa = 10.6AEE48 pKa = 4.47HH49 pKa = 6.07EE50 pKa = 4.33TAKK53 pKa = 10.91LGTNLFMVRR62 pKa = 11.84SGQLSFRR69 pKa = 11.84PSGSSRR75 pKa = 11.84VRR77 pKa = 11.84GLARR81 pKa = 11.84NFTRR85 pKa = 11.84QDD87 pKa = 3.02AKK89 pKa = 11.36ALLAGVPAARR99 pKa = 11.84AGAPFVLATSPIRR112 pKa = 11.84SGDD115 pKa = 3.65LQINCQLVATLPSYY129 pKa = 10.02ATVRR133 pKa = 11.84NVKK136 pKa = 9.83AAWGRR141 pKa = 11.84FLSASDD147 pKa = 3.19EE148 pKa = 4.18AEE150 pKa = 3.93RR151 pKa = 11.84AKK153 pKa = 11.0VCVLGASIARR163 pKa = 11.84RR164 pKa = 11.84LFGDD168 pKa = 3.39GQGAVGQQVMLHH180 pKa = 5.86RR181 pKa = 11.84AGLRR185 pKa = 11.84VIGVMEE191 pKa = 4.53PKK193 pKa = 10.54GADD196 pKa = 2.97IAGTDD201 pKa = 3.01QDD203 pKa = 3.78EE204 pKa = 4.55QVFVPLTTYY213 pKa = 8.62MRR215 pKa = 11.84RR216 pKa = 11.84LANQDD221 pKa = 3.6WISGVYY227 pKa = 9.66LQLADD232 pKa = 3.53SQAIEE237 pKa = 4.27EE238 pKa = 4.36ARR240 pKa = 11.84QAAATILRR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84HH251 pKa = 6.01SIDD254 pKa = 3.12VGEE257 pKa = 5.32DD258 pKa = 3.24DD259 pKa = 6.65DD260 pKa = 4.47FTLLAAQDD268 pKa = 3.61TMRR271 pKa = 11.84VQRR274 pKa = 11.84DD275 pKa = 3.31ALALVQTLGLLSSSISFAVGGLGILSIMVLLVRR308 pKa = 11.84TRR310 pKa = 11.84RR311 pKa = 11.84LEE313 pKa = 3.78IGVRR317 pKa = 11.84RR318 pKa = 11.84AVGAKK323 pKa = 9.67RR324 pKa = 11.84RR325 pKa = 11.84DD326 pKa = 3.27IVRR329 pKa = 11.84QFMFEE334 pKa = 3.94AGLMAGIGGGLGVLSSLLLVLVVCRR359 pKa = 11.84IGEE362 pKa = 4.06MPLAYY367 pKa = 10.26DD368 pKa = 3.6PVLIGSSLFGSLLLGLAAGAYY389 pKa = 7.53PAWQAANVEE398 pKa = 4.14ILQVLRR404 pKa = 11.84NEE406 pKa = 4.23
MM1 pKa = 7.3FFSLKK6 pKa = 9.98IALSSLRR13 pKa = 11.84AHH15 pKa = 7.57RR16 pKa = 11.84LRR18 pKa = 11.84SILAMVGVFLGALALTGVQHH38 pKa = 6.5VSRR41 pKa = 11.84SMLLKK46 pKa = 10.6AEE48 pKa = 4.47HH49 pKa = 6.07EE50 pKa = 4.33TAKK53 pKa = 10.91LGTNLFMVRR62 pKa = 11.84SGQLSFRR69 pKa = 11.84PSGSSRR75 pKa = 11.84VRR77 pKa = 11.84GLARR81 pKa = 11.84NFTRR85 pKa = 11.84QDD87 pKa = 3.02AKK89 pKa = 11.36ALLAGVPAARR99 pKa = 11.84AGAPFVLATSPIRR112 pKa = 11.84SGDD115 pKa = 3.65LQINCQLVATLPSYY129 pKa = 10.02ATVRR133 pKa = 11.84NVKK136 pKa = 9.83AAWGRR141 pKa = 11.84FLSASDD147 pKa = 3.19EE148 pKa = 4.18AEE150 pKa = 3.93RR151 pKa = 11.84AKK153 pKa = 11.0VCVLGASIARR163 pKa = 11.84RR164 pKa = 11.84LFGDD168 pKa = 3.39GQGAVGQQVMLHH180 pKa = 5.86RR181 pKa = 11.84AGLRR185 pKa = 11.84VIGVMEE191 pKa = 4.53PKK193 pKa = 10.54GADD196 pKa = 2.97IAGTDD201 pKa = 3.01QDD203 pKa = 3.78EE204 pKa = 4.55QVFVPLTTYY213 pKa = 8.62MRR215 pKa = 11.84RR216 pKa = 11.84LANQDD221 pKa = 3.6WISGVYY227 pKa = 9.66LQLADD232 pKa = 3.53SQAIEE237 pKa = 4.27EE238 pKa = 4.36ARR240 pKa = 11.84QAAATILRR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84HH251 pKa = 6.01SIDD254 pKa = 3.12VGEE257 pKa = 5.32DD258 pKa = 3.24DD259 pKa = 6.65DD260 pKa = 4.47FTLLAAQDD268 pKa = 3.61TMRR271 pKa = 11.84VQRR274 pKa = 11.84DD275 pKa = 3.31ALALVQTLGLLSSSISFAVGGLGILSIMVLLVRR308 pKa = 11.84TRR310 pKa = 11.84RR311 pKa = 11.84LEE313 pKa = 3.78IGVRR317 pKa = 11.84RR318 pKa = 11.84AVGAKK323 pKa = 9.67RR324 pKa = 11.84RR325 pKa = 11.84DD326 pKa = 3.27IVRR329 pKa = 11.84QFMFEE334 pKa = 3.94AGLMAGIGGGLGVLSSLLLVLVVCRR359 pKa = 11.84IGEE362 pKa = 4.06MPLAYY367 pKa = 10.26DD368 pKa = 3.6PVLIGSSLFGSLLLGLAAGAYY389 pKa = 7.53PAWQAANVEE398 pKa = 4.14ILQVLRR404 pKa = 11.84NEE406 pKa = 4.23
Molecular weight: 43.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
882519 |
33 |
1929 |
319.5 |
35.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.283 ± 0.053 | 1.322 ± 0.021 |
5.411 ± 0.035 | 6.661 ± 0.047 |
3.972 ± 0.031 | 7.79 ± 0.038 |
2.081 ± 0.02 | 5.619 ± 0.041 |
4.459 ± 0.039 | 11.513 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.309 ± 0.022 | 3.127 ± 0.026 |
4.515 ± 0.029 | 4.253 ± 0.031 |
6.328 ± 0.043 | 5.692 ± 0.033 |
4.823 ± 0.029 | 7.145 ± 0.039 |
1.056 ± 0.017 | 2.641 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
