
Pea enation mosaic virus-1 (strain WSG) (PEMV-1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Enamovirus; Pea enation mosaic virus 1
Average proteome isoelectric point is 7.64
Get precalculated fractions of proteins
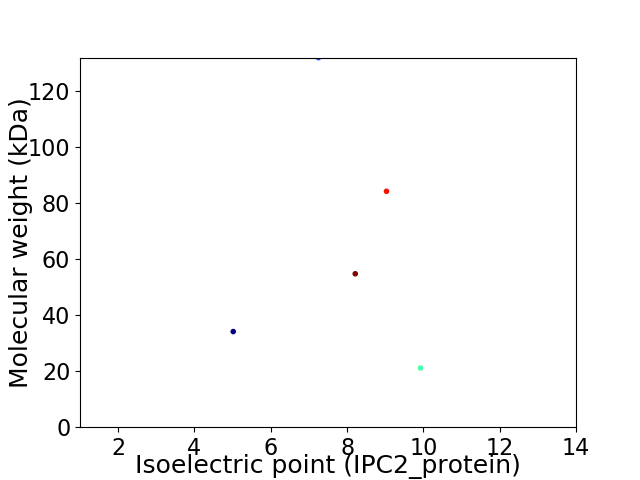
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
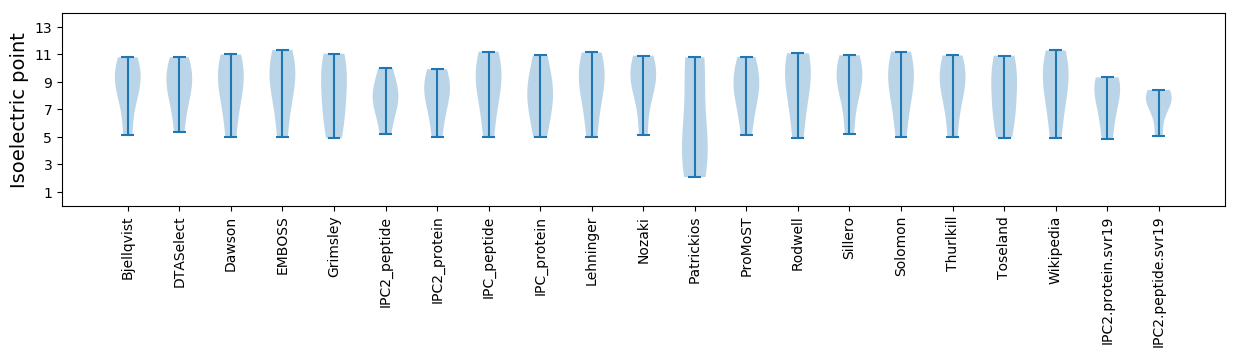
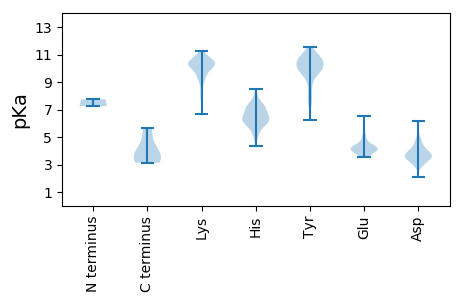
Protein with the lowest isoelectric point:
>sp|Q84710|P1_PEMVW Protein P1 OS=Pea enation mosaic virus-1 (strain WSG) OX=693989 GN=ORF1 PE=1 SV=1
MM1 pKa = 7.47HH2 pKa = 7.61GIEE5 pKa = 4.54QPQLPLDD12 pKa = 3.88YY13 pKa = 10.96VHH15 pKa = 7.27RR16 pKa = 11.84CASTSFLLASLDD28 pKa = 3.7GLLSEE33 pKa = 4.94ARR35 pKa = 11.84EE36 pKa = 4.07LSGPLALITSSYY48 pKa = 9.67YY49 pKa = 11.01LLVSIALCWAIPGSFWYY66 pKa = 10.31RR67 pKa = 11.84PGCWLQPVSGRR78 pKa = 11.84NLIFCGPTEE87 pKa = 3.89ALQRR91 pKa = 11.84FRR93 pKa = 11.84LYY95 pKa = 10.64AARR98 pKa = 11.84LGLVLSEE105 pKa = 3.75NCPRR109 pKa = 11.84HH110 pKa = 5.4GQSAAITLQSYY121 pKa = 7.64WALPNNIWMDD131 pKa = 3.77MAQLDD136 pKa = 4.73LLTFSMPIANTFAYY150 pKa = 10.45LADD153 pKa = 4.07CEE155 pKa = 4.92ARR157 pKa = 11.84FPPIVEE163 pKa = 4.53GVGSAYY169 pKa = 10.26YY170 pKa = 10.78VPTLLGLTHH179 pKa = 6.64QDD181 pKa = 2.8PRR183 pKa = 11.84LYY185 pKa = 10.65LALRR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84NLDD194 pKa = 3.24LSGEE198 pKa = 4.13PHH200 pKa = 6.56RR201 pKa = 11.84VRR203 pKa = 11.84PGVLEE208 pKa = 4.57SMALLCSSVRR218 pKa = 11.84STSRR222 pKa = 11.84SRR224 pKa = 11.84QIPPLYY230 pKa = 10.31GSVLHH235 pKa = 6.58HH236 pKa = 6.11VLGLAEE242 pKa = 4.39RR243 pKa = 11.84DD244 pKa = 3.94CILFDD249 pKa = 3.51TDD251 pKa = 4.27SNYY254 pKa = 10.45SSYY257 pKa = 7.4THH259 pKa = 7.05RR260 pKa = 11.84VLEE263 pKa = 3.99QDD265 pKa = 3.89RR266 pKa = 11.84NRR268 pKa = 11.84ADD270 pKa = 3.13QSLFSIDD277 pKa = 3.72LEE279 pKa = 4.42YY280 pKa = 11.55VHH282 pKa = 7.37DD283 pKa = 5.02LEE285 pKa = 6.37LIALGYY291 pKa = 10.13SDD293 pKa = 6.09EE294 pKa = 5.43DD295 pKa = 6.14DD296 pKa = 4.02EE297 pKa = 6.55DD298 pKa = 5.02LDD300 pKa = 4.49NFFF303 pKa = 5.65
MM1 pKa = 7.47HH2 pKa = 7.61GIEE5 pKa = 4.54QPQLPLDD12 pKa = 3.88YY13 pKa = 10.96VHH15 pKa = 7.27RR16 pKa = 11.84CASTSFLLASLDD28 pKa = 3.7GLLSEE33 pKa = 4.94ARR35 pKa = 11.84EE36 pKa = 4.07LSGPLALITSSYY48 pKa = 9.67YY49 pKa = 11.01LLVSIALCWAIPGSFWYY66 pKa = 10.31RR67 pKa = 11.84PGCWLQPVSGRR78 pKa = 11.84NLIFCGPTEE87 pKa = 3.89ALQRR91 pKa = 11.84FRR93 pKa = 11.84LYY95 pKa = 10.64AARR98 pKa = 11.84LGLVLSEE105 pKa = 3.75NCPRR109 pKa = 11.84HH110 pKa = 5.4GQSAAITLQSYY121 pKa = 7.64WALPNNIWMDD131 pKa = 3.77MAQLDD136 pKa = 4.73LLTFSMPIANTFAYY150 pKa = 10.45LADD153 pKa = 4.07CEE155 pKa = 4.92ARR157 pKa = 11.84FPPIVEE163 pKa = 4.53GVGSAYY169 pKa = 10.26YY170 pKa = 10.78VPTLLGLTHH179 pKa = 6.64QDD181 pKa = 2.8PRR183 pKa = 11.84LYY185 pKa = 10.65LALRR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84NLDD194 pKa = 3.24LSGEE198 pKa = 4.13PHH200 pKa = 6.56RR201 pKa = 11.84VRR203 pKa = 11.84PGVLEE208 pKa = 4.57SMALLCSSVRR218 pKa = 11.84STSRR222 pKa = 11.84SRR224 pKa = 11.84QIPPLYY230 pKa = 10.31GSVLHH235 pKa = 6.58HH236 pKa = 6.11VLGLAEE242 pKa = 4.39RR243 pKa = 11.84DD244 pKa = 3.94CILFDD249 pKa = 3.51TDD251 pKa = 4.27SNYY254 pKa = 10.45SSYY257 pKa = 7.4THH259 pKa = 7.05RR260 pKa = 11.84VLEE263 pKa = 3.99QDD265 pKa = 3.89RR266 pKa = 11.84NRR268 pKa = 11.84ADD270 pKa = 3.13QSLFSIDD277 pKa = 3.72LEE279 pKa = 4.42YY280 pKa = 11.55VHH282 pKa = 7.37DD283 pKa = 5.02LEE285 pKa = 6.37LIALGYY291 pKa = 10.13SDD293 pKa = 6.09EE294 pKa = 5.43DD295 pKa = 6.14DD296 pKa = 4.02EE297 pKa = 6.55DD298 pKa = 5.02LDD300 pKa = 4.49NFFF303 pKa = 5.65
Molecular weight: 34.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P29154|RDRP_PEMVW Protein P1-P2 OS=Pea enation mosaic virus-1 (strain WSG) OX=693989 GN=ORF1/ORF2 PE=3 SV=2
MM1 pKa = 7.25PTRR4 pKa = 11.84SRR6 pKa = 11.84SKK8 pKa = 9.39ANQRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84VVVVAPSMAQPRR30 pKa = 11.84TQSRR34 pKa = 11.84RR35 pKa = 11.84PRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84NKK41 pKa = 10.39RR42 pKa = 11.84GGGLNGSHH50 pKa = 5.93TVDD53 pKa = 4.09FSMVHH58 pKa = 6.25GPFNGNATGTVKK70 pKa = 10.84FGPSSDD76 pKa = 3.84CQCIKK81 pKa = 11.1GNLAAYY87 pKa = 8.97QKK89 pKa = 11.0YY90 pKa = 9.83RR91 pKa = 11.84IVWLKK96 pKa = 10.59VVYY99 pKa = 10.23QSEE102 pKa = 4.02AAATDD107 pKa = 3.93RR108 pKa = 11.84GCIAYY113 pKa = 9.48HH114 pKa = 6.44VDD116 pKa = 2.87TSTTKK121 pKa = 10.26KK122 pKa = 10.46AADD125 pKa = 3.62VVLLDD130 pKa = 3.38TWNIRR135 pKa = 11.84SNGSATFGRR144 pKa = 11.84EE145 pKa = 3.72ILGDD149 pKa = 3.82QPWYY153 pKa = 10.36EE154 pKa = 4.24SNKK157 pKa = 10.14DD158 pKa = 3.31QFFFLYY164 pKa = 9.98RR165 pKa = 11.84GTGGTDD171 pKa = 2.94VAGHH175 pKa = 5.26YY176 pKa = 9.54RR177 pKa = 11.84ISGRR181 pKa = 11.84IQLMNASLL189 pKa = 3.54
MM1 pKa = 7.25PTRR4 pKa = 11.84SRR6 pKa = 11.84SKK8 pKa = 9.39ANQRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84VVVVAPSMAQPRR30 pKa = 11.84TQSRR34 pKa = 11.84RR35 pKa = 11.84PRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84NKK41 pKa = 10.39RR42 pKa = 11.84GGGLNGSHH50 pKa = 5.93TVDD53 pKa = 4.09FSMVHH58 pKa = 6.25GPFNGNATGTVKK70 pKa = 10.84FGPSSDD76 pKa = 3.84CQCIKK81 pKa = 11.1GNLAAYY87 pKa = 8.97QKK89 pKa = 11.0YY90 pKa = 9.83RR91 pKa = 11.84IVWLKK96 pKa = 10.59VVYY99 pKa = 10.23QSEE102 pKa = 4.02AAATDD107 pKa = 3.93RR108 pKa = 11.84GCIAYY113 pKa = 9.48HH114 pKa = 6.44VDD116 pKa = 2.87TSTTKK121 pKa = 10.26KK122 pKa = 10.46AADD125 pKa = 3.62VVLLDD130 pKa = 3.38TWNIRR135 pKa = 11.84SNGSATFGRR144 pKa = 11.84EE145 pKa = 3.72ILGDD149 pKa = 3.82QPWYY153 pKa = 10.36EE154 pKa = 4.24SNKK157 pKa = 10.14DD158 pKa = 3.31QFFFLYY164 pKa = 9.98RR165 pKa = 11.84GTGGTDD171 pKa = 2.94VAGHH175 pKa = 5.26YY176 pKa = 9.54RR177 pKa = 11.84ISGRR181 pKa = 11.84IQLMNASLL189 pKa = 3.54
Molecular weight: 21.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2926 |
189 |
1181 |
585.2 |
65.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.972 ± 0.232 | 2.016 ± 0.143 |
4.99 ± 0.359 | 4.887 ± 0.561 |
3.691 ± 0.246 | 7.04 ± 0.589 |
1.777 ± 0.196 | 4.375 ± 0.132 |
5.195 ± 0.745 | 9.877 ± 1.365 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.085 ± 0.174 | 3.008 ± 0.368 |
6.528 ± 0.547 | 4.17 ± 0.114 |
6.699 ± 0.735 | 9.364 ± 0.193 |
6.425 ± 0.42 | 5.742 ± 0.277 |
2.495 ± 0.189 | 2.666 ± 0.538 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
