
Shayang Fly Virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alphadrosrhavirus; Shayang alphadrosrhavirus
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
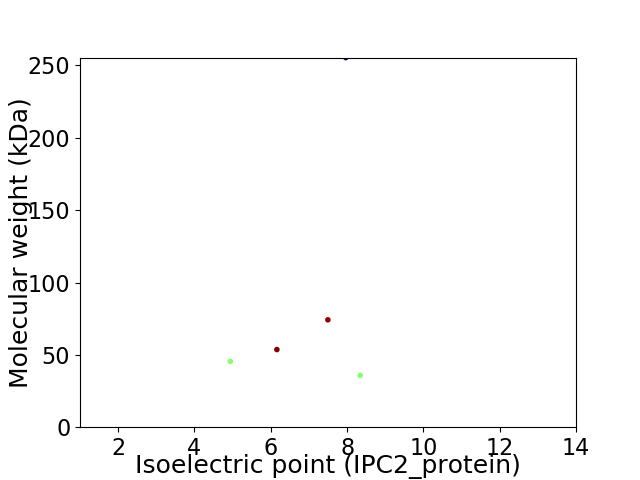
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KXF0|A0A0B5KXF0_9RHAB ORF2 OS=Shayang Fly Virus 3 OX=1608067 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.36SFSISRR7 pKa = 11.84QICQLEE13 pKa = 4.23NCIHH17 pKa = 6.44GPEE20 pKa = 4.01EE21 pKa = 4.05HH22 pKa = 7.28PISVDD27 pKa = 3.28CMEE30 pKa = 4.98DD31 pKa = 3.42QNEE34 pKa = 4.13EE35 pKa = 4.0EE36 pKa = 5.22LLNLDD41 pKa = 4.86PDD43 pKa = 4.02DD44 pKa = 5.31QAILDD49 pKa = 3.6NAQFLKK55 pKa = 10.76ASSHH59 pKa = 7.06FYY61 pKa = 9.96GQEE64 pKa = 3.73DD65 pKa = 3.77ATGKK69 pKa = 9.98IEE71 pKa = 3.95TLLRR75 pKa = 11.84SDD77 pKa = 4.54SLDD80 pKa = 3.77DD81 pKa = 3.88NFAAYY86 pKa = 10.41VNSSNEE92 pKa = 4.13DD93 pKa = 3.32KK94 pKa = 11.17DD95 pKa = 3.92KK96 pKa = 11.55EE97 pKa = 4.41NYY99 pKa = 9.82NPDD102 pKa = 3.21VNQNDD107 pKa = 3.57SLLRR111 pKa = 11.84KK112 pKa = 9.78SEE114 pKa = 3.76GGEE117 pKa = 5.1DD118 pKa = 2.99INQAINNFYY127 pKa = 10.61QYY129 pKa = 9.58WSKK132 pKa = 11.26PEE134 pKa = 3.92NQEE137 pKa = 3.67ILYY140 pKa = 7.15KK141 pKa = 10.44TPLIVFDD148 pKa = 5.56SIIKK152 pKa = 10.21DD153 pKa = 3.38PRR155 pKa = 11.84FEE157 pKa = 4.56NYY159 pKa = 10.31CNMFSSCRR167 pKa = 11.84LKK169 pKa = 10.13FHH171 pKa = 6.05QLGVEE176 pKa = 4.39RR177 pKa = 11.84GRR179 pKa = 11.84DD180 pKa = 3.59LMIHH184 pKa = 6.85FISEE188 pKa = 3.69RR189 pKa = 11.84NIRR192 pKa = 11.84YY193 pKa = 7.62FQNFDD198 pKa = 2.83IMLFGANTVSQTANQKK214 pKa = 10.35DD215 pKa = 3.92LEE217 pKa = 4.21LLKK220 pKa = 10.98VEE222 pKa = 4.85FEE224 pKa = 4.68SKK226 pKa = 10.7VSFLVDD232 pKa = 3.29SVKK235 pKa = 10.05TVEE238 pKa = 4.08EE239 pKa = 4.44HH240 pKa = 6.6IKK242 pKa = 10.12TIDD245 pKa = 3.48SSHH248 pKa = 6.11EE249 pKa = 3.85ALRR252 pKa = 11.84DD253 pKa = 3.65TINCVSEE260 pKa = 4.11AAHH263 pKa = 6.15GLSAAKK269 pKa = 10.13RR270 pKa = 11.84SFEE273 pKa = 4.06LLQEE277 pKa = 4.42RR278 pKa = 11.84DD279 pKa = 3.36PSYY282 pKa = 11.66SPVISNTARR291 pKa = 11.84DD292 pKa = 3.94MPKK295 pKa = 10.28KK296 pKa = 10.28SVSIPKK302 pKa = 10.33LKK304 pKa = 9.4TVYY307 pKa = 10.21KK308 pKa = 10.04KK309 pKa = 10.11EE310 pKa = 4.04HH311 pKa = 6.0LSIPIGMNDD320 pKa = 3.93EE321 pKa = 4.58LVISQISVYY330 pKa = 10.99NKK332 pKa = 10.34GLTLSDD338 pKa = 4.0FKK340 pKa = 11.37LFEE343 pKa = 4.72GLSKK347 pKa = 10.44MKK349 pKa = 10.94AEE351 pKa = 4.19VFLRR355 pKa = 11.84ALNLPWALLTKK366 pKa = 9.2WANSSPDD373 pKa = 3.23NSSIGIARR381 pKa = 11.84TLIPMIMNLVPLPAKK396 pKa = 10.83VEE398 pKa = 4.27FF399 pKa = 4.45
MM1 pKa = 7.36SFSISRR7 pKa = 11.84QICQLEE13 pKa = 4.23NCIHH17 pKa = 6.44GPEE20 pKa = 4.01EE21 pKa = 4.05HH22 pKa = 7.28PISVDD27 pKa = 3.28CMEE30 pKa = 4.98DD31 pKa = 3.42QNEE34 pKa = 4.13EE35 pKa = 4.0EE36 pKa = 5.22LLNLDD41 pKa = 4.86PDD43 pKa = 4.02DD44 pKa = 5.31QAILDD49 pKa = 3.6NAQFLKK55 pKa = 10.76ASSHH59 pKa = 7.06FYY61 pKa = 9.96GQEE64 pKa = 3.73DD65 pKa = 3.77ATGKK69 pKa = 9.98IEE71 pKa = 3.95TLLRR75 pKa = 11.84SDD77 pKa = 4.54SLDD80 pKa = 3.77DD81 pKa = 3.88NFAAYY86 pKa = 10.41VNSSNEE92 pKa = 4.13DD93 pKa = 3.32KK94 pKa = 11.17DD95 pKa = 3.92KK96 pKa = 11.55EE97 pKa = 4.41NYY99 pKa = 9.82NPDD102 pKa = 3.21VNQNDD107 pKa = 3.57SLLRR111 pKa = 11.84KK112 pKa = 9.78SEE114 pKa = 3.76GGEE117 pKa = 5.1DD118 pKa = 2.99INQAINNFYY127 pKa = 10.61QYY129 pKa = 9.58WSKK132 pKa = 11.26PEE134 pKa = 3.92NQEE137 pKa = 3.67ILYY140 pKa = 7.15KK141 pKa = 10.44TPLIVFDD148 pKa = 5.56SIIKK152 pKa = 10.21DD153 pKa = 3.38PRR155 pKa = 11.84FEE157 pKa = 4.56NYY159 pKa = 10.31CNMFSSCRR167 pKa = 11.84LKK169 pKa = 10.13FHH171 pKa = 6.05QLGVEE176 pKa = 4.39RR177 pKa = 11.84GRR179 pKa = 11.84DD180 pKa = 3.59LMIHH184 pKa = 6.85FISEE188 pKa = 3.69RR189 pKa = 11.84NIRR192 pKa = 11.84YY193 pKa = 7.62FQNFDD198 pKa = 2.83IMLFGANTVSQTANQKK214 pKa = 10.35DD215 pKa = 3.92LEE217 pKa = 4.21LLKK220 pKa = 10.98VEE222 pKa = 4.85FEE224 pKa = 4.68SKK226 pKa = 10.7VSFLVDD232 pKa = 3.29SVKK235 pKa = 10.05TVEE238 pKa = 4.08EE239 pKa = 4.44HH240 pKa = 6.6IKK242 pKa = 10.12TIDD245 pKa = 3.48SSHH248 pKa = 6.11EE249 pKa = 3.85ALRR252 pKa = 11.84DD253 pKa = 3.65TINCVSEE260 pKa = 4.11AAHH263 pKa = 6.15GLSAAKK269 pKa = 10.13RR270 pKa = 11.84SFEE273 pKa = 4.06LLQEE277 pKa = 4.42RR278 pKa = 11.84DD279 pKa = 3.36PSYY282 pKa = 11.66SPVISNTARR291 pKa = 11.84DD292 pKa = 3.94MPKK295 pKa = 10.28KK296 pKa = 10.28SVSIPKK302 pKa = 10.33LKK304 pKa = 9.4TVYY307 pKa = 10.21KK308 pKa = 10.04KK309 pKa = 10.11EE310 pKa = 4.04HH311 pKa = 6.0LSIPIGMNDD320 pKa = 3.93EE321 pKa = 4.58LVISQISVYY330 pKa = 10.99NKK332 pKa = 10.34GLTLSDD338 pKa = 4.0FKK340 pKa = 11.37LFEE343 pKa = 4.72GLSKK347 pKa = 10.44MKK349 pKa = 10.94AEE351 pKa = 4.19VFLRR355 pKa = 11.84ALNLPWALLTKK366 pKa = 9.2WANSSPDD373 pKa = 3.23NSSIGIARR381 pKa = 11.84TLIPMIMNLVPLPAKK396 pKa = 10.83VEE398 pKa = 4.27FF399 pKa = 4.45
Molecular weight: 45.47 kDa
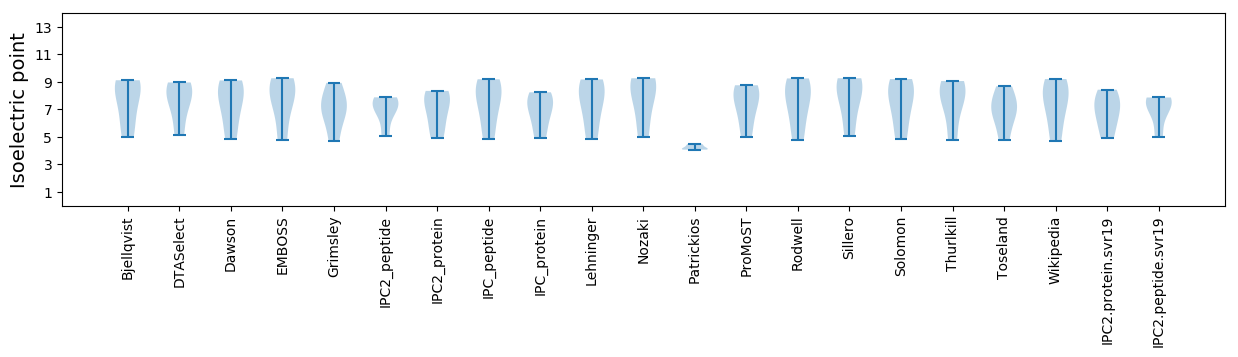
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KK44|A0A0B5KK44_9RHAB Glycoprotein OS=Shayang Fly Virus 3 OX=1608067 GN=G PE=4 SV=1
MM1 pKa = 7.62LGLFSTSSRR10 pKa = 11.84PISPISFCNLNSRR23 pKa = 11.84SLSSIKK29 pKa = 9.75TIYY32 pKa = 7.5FTPSSSSFSTMTEE45 pKa = 3.75PSTSKK50 pKa = 10.95SLVTSIPHH58 pKa = 5.49YY59 pKa = 10.99SPLQTEE65 pKa = 4.45QISLVIKK72 pKa = 10.21GAIDD76 pKa = 3.47VSGPKK81 pKa = 9.94LDD83 pKa = 3.66FVEE86 pKa = 4.81FLWRR90 pKa = 11.84VCCVLAYY97 pKa = 8.81EE98 pKa = 4.27ATRR101 pKa = 11.84LKK103 pKa = 10.69SVSRR107 pKa = 11.84NTQYY111 pKa = 11.41LVASILYY118 pKa = 9.27EE119 pKa = 3.97EE120 pKa = 5.08LRR122 pKa = 11.84SSSYY126 pKa = 11.11LGTFSSPDD134 pKa = 3.11IGPTCEE140 pKa = 4.2LLCPIEE146 pKa = 4.28VKK148 pKa = 10.78GFTEE152 pKa = 5.04FPIEE156 pKa = 4.22SSSFPQIRR164 pKa = 11.84KK165 pKa = 8.67SLNRR169 pKa = 11.84VFRR172 pKa = 11.84YY173 pKa = 8.26QVKK176 pKa = 9.41PGSILRR182 pKa = 11.84IRR184 pKa = 11.84AGLTIQACKK193 pKa = 10.53FNDD196 pKa = 3.63KK197 pKa = 9.7YY198 pKa = 11.41RR199 pKa = 11.84EE200 pKa = 3.67MARR203 pKa = 11.84IKK205 pKa = 10.6GYY207 pKa = 10.94VPLLPPSNEE216 pKa = 3.59HH217 pKa = 6.45LISYY221 pKa = 9.35FSLLEE226 pKa = 3.91NSIEE230 pKa = 4.09TAGAYY235 pKa = 9.1NEE237 pKa = 4.35KK238 pKa = 10.51DD239 pKa = 2.82ISSFIASGIDD249 pKa = 3.15ALAEE253 pKa = 4.1KK254 pKa = 10.16EE255 pKa = 4.04ADD257 pKa = 3.7PPLSFIKK264 pKa = 10.75LIRR267 pKa = 11.84DD268 pKa = 3.66KK269 pKa = 10.81IFKK272 pKa = 9.96QNPPASIGPGKK283 pKa = 10.29ASSGTKK289 pKa = 10.08PPDD292 pKa = 3.28GLRR295 pKa = 11.84ILTQSKK301 pKa = 10.72LKK303 pKa = 10.32GSKK306 pKa = 10.11YY307 pKa = 10.75DD308 pKa = 3.66LALCEE313 pKa = 4.33STSSIKK319 pKa = 10.41KK320 pKa = 10.0SEE322 pKa = 4.39SINN325 pKa = 3.46
MM1 pKa = 7.62LGLFSTSSRR10 pKa = 11.84PISPISFCNLNSRR23 pKa = 11.84SLSSIKK29 pKa = 9.75TIYY32 pKa = 7.5FTPSSSSFSTMTEE45 pKa = 3.75PSTSKK50 pKa = 10.95SLVTSIPHH58 pKa = 5.49YY59 pKa = 10.99SPLQTEE65 pKa = 4.45QISLVIKK72 pKa = 10.21GAIDD76 pKa = 3.47VSGPKK81 pKa = 9.94LDD83 pKa = 3.66FVEE86 pKa = 4.81FLWRR90 pKa = 11.84VCCVLAYY97 pKa = 8.81EE98 pKa = 4.27ATRR101 pKa = 11.84LKK103 pKa = 10.69SVSRR107 pKa = 11.84NTQYY111 pKa = 11.41LVASILYY118 pKa = 9.27EE119 pKa = 3.97EE120 pKa = 5.08LRR122 pKa = 11.84SSSYY126 pKa = 11.11LGTFSSPDD134 pKa = 3.11IGPTCEE140 pKa = 4.2LLCPIEE146 pKa = 4.28VKK148 pKa = 10.78GFTEE152 pKa = 5.04FPIEE156 pKa = 4.22SSSFPQIRR164 pKa = 11.84KK165 pKa = 8.67SLNRR169 pKa = 11.84VFRR172 pKa = 11.84YY173 pKa = 8.26QVKK176 pKa = 9.41PGSILRR182 pKa = 11.84IRR184 pKa = 11.84AGLTIQACKK193 pKa = 10.53FNDD196 pKa = 3.63KK197 pKa = 9.7YY198 pKa = 11.41RR199 pKa = 11.84EE200 pKa = 3.67MARR203 pKa = 11.84IKK205 pKa = 10.6GYY207 pKa = 10.94VPLLPPSNEE216 pKa = 3.59HH217 pKa = 6.45LISYY221 pKa = 9.35FSLLEE226 pKa = 3.91NSIEE230 pKa = 4.09TAGAYY235 pKa = 9.1NEE237 pKa = 4.35KK238 pKa = 10.51DD239 pKa = 2.82ISSFIASGIDD249 pKa = 3.15ALAEE253 pKa = 4.1KK254 pKa = 10.16EE255 pKa = 4.04ADD257 pKa = 3.7PPLSFIKK264 pKa = 10.75LIRR267 pKa = 11.84DD268 pKa = 3.66KK269 pKa = 10.81IFKK272 pKa = 9.96QNPPASIGPGKK283 pKa = 10.29ASSGTKK289 pKa = 10.08PPDD292 pKa = 3.28GLRR295 pKa = 11.84ILTQSKK301 pKa = 10.72LKK303 pKa = 10.32GSKK306 pKa = 10.11YY307 pKa = 10.75DD308 pKa = 3.66LALCEE313 pKa = 4.33STSSIKK319 pKa = 10.41KK320 pKa = 10.0SEE322 pKa = 4.39SINN325 pKa = 3.46
Molecular weight: 35.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4082 |
325 |
2231 |
816.4 |
92.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.532 ± 0.733 | 1.788 ± 0.052 |
4.998 ± 0.335 | 5.194 ± 0.458 |
5.781 ± 0.42 | 4.483 ± 0.308 |
2.327 ± 0.305 | 8.305 ± 0.483 |
6.835 ± 0.261 | 10.828 ± 0.502 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.597 ± 0.301 | 5.512 ± 0.329 |
4.483 ± 0.321 | 3.503 ± 0.276 |
3.944 ± 0.246 | 10.289 ± 0.764 |
5.267 ± 0.395 | 4.532 ± 0.349 |
1.249 ± 0.206 | 3.552 ± 0.206 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
