
Cronartium ribicola mitovirus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 7.72
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A191KCP3|A0A191KCP3_9VIRU RNA-dependent RNA polymerase OS=Cronartium ribicola mitovirus 3 OX=1816486 GN=RdRp PE=4 SV=1
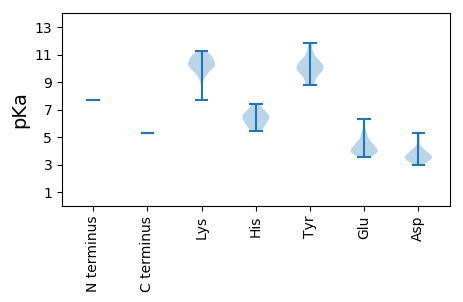
MM1 pKa = 7.7NMNQNKK7 pKa = 9.85AYY9 pKa = 9.89SALVQAIFTFLIYY22 pKa = 9.81YY23 pKa = 10.23SPGFEE28 pKa = 3.92SDD30 pKa = 3.6KK31 pKa = 10.99IVIILSDD38 pKa = 4.0FKK40 pKa = 10.88RR41 pKa = 11.84HH42 pKa = 5.88CDD44 pKa = 3.32QRR46 pKa = 11.84GIKK49 pKa = 10.25AGISWIKK56 pKa = 10.27DD57 pKa = 3.0LRR59 pKa = 11.84LCFTKK64 pKa = 10.93YY65 pKa = 8.91QAGDD69 pKa = 3.59SKK71 pKa = 11.22PKK73 pKa = 10.81VNIKK77 pKa = 9.57CHH79 pKa = 6.27PNGCPRR85 pKa = 11.84LIWDD89 pKa = 3.78NYY91 pKa = 10.41KK92 pKa = 10.88DD93 pKa = 3.72VLNDD97 pKa = 3.52PKK99 pKa = 11.04GIGIIITLLSISRR112 pKa = 11.84PFEE115 pKa = 4.57GIFEE119 pKa = 4.66PSLSDD124 pKa = 3.18ITDD127 pKa = 3.59TPKK130 pKa = 10.55TDD132 pKa = 3.44LGIIKK137 pKa = 10.03DD138 pKa = 3.9LEE140 pKa = 4.17EE141 pKa = 3.9FLPAFFEE148 pKa = 4.61EE149 pKa = 4.95VVHH152 pKa = 5.84VRR154 pKa = 11.84KK155 pKa = 10.07EE156 pKa = 3.89SGQLKK161 pKa = 8.95RR162 pKa = 11.84PKK164 pKa = 9.39WSSWLNNIKK173 pKa = 10.07IAPNGVSINSSYY185 pKa = 11.81ADD187 pKa = 3.48LDD189 pKa = 3.72LHH191 pKa = 7.4NEE193 pKa = 4.04EE194 pKa = 5.2IIRR197 pKa = 11.84DD198 pKa = 3.91LFEE201 pKa = 6.35LGGDD205 pKa = 3.96CFEE208 pKa = 5.42KK209 pKa = 10.34EE210 pKa = 3.91YY211 pKa = 11.46SSIKK215 pKa = 10.47DD216 pKa = 3.29YY217 pKa = 11.04WKK219 pKa = 10.73DD220 pKa = 3.36PEE222 pKa = 4.33GQKK225 pKa = 10.58EE226 pKa = 3.86ITKK229 pKa = 10.31NYY231 pKa = 9.43PIVNTYY237 pKa = 8.83KK238 pKa = 10.41SKK240 pKa = 10.33YY241 pKa = 8.94LGRR244 pKa = 11.84LAVIPSPEE252 pKa = 3.98GKK254 pKa = 9.96SRR256 pKa = 11.84VIAIGDD262 pKa = 3.5YY263 pKa = 9.59WSQNVLFPLHH273 pKa = 6.31NKK275 pKa = 9.41IFGLLKK281 pKa = 10.49RR282 pKa = 11.84IPQDD286 pKa = 3.04LTFAQEE292 pKa = 3.88QGLSKK297 pKa = 10.25IKK299 pKa = 10.12KK300 pKa = 9.97DD301 pKa = 3.43RR302 pKa = 11.84DD303 pKa = 3.19HH304 pKa = 7.15SYY306 pKa = 10.45WSLDD310 pKa = 3.44LKK312 pKa = 11.0SATDD316 pKa = 3.74RR317 pKa = 11.84FPIEE321 pKa = 3.66LQRR324 pKa = 11.84RR325 pKa = 11.84IISYY329 pKa = 9.95IYY331 pKa = 9.91GEE333 pKa = 4.59SYY335 pKa = 10.56SISWRR340 pKa = 11.84NLMSRR345 pKa = 11.84PFYY348 pKa = 9.87QTISGKK354 pKa = 9.86SEE356 pKa = 3.57VTYY359 pKa = 10.61GAGQPIGIYY368 pKa = 10.05SSWSVFTLCHH378 pKa = 6.5HH379 pKa = 6.57FVLYY383 pKa = 10.1CAQRR387 pKa = 11.84RR388 pKa = 11.84SIALKK393 pKa = 9.35KK394 pKa = 9.92GRR396 pKa = 11.84YY397 pKa = 8.78VILGDD402 pKa = 5.28DD403 pKa = 4.11IVISDD408 pKa = 4.43DD409 pKa = 3.6NLAIEE414 pKa = 4.24YY415 pKa = 9.8KK416 pKa = 10.65KK417 pKa = 10.8LINLLGVEE425 pKa = 3.93ISEE428 pKa = 4.5HH429 pKa = 5.4KK430 pKa = 7.73THH432 pKa = 6.93KK433 pKa = 10.26SKK435 pKa = 10.85HH436 pKa = 5.69SYY438 pKa = 10.09EE439 pKa = 3.78IAKK442 pKa = 10.03RR443 pKa = 11.84WITEE447 pKa = 3.86KK448 pKa = 11.04GEE450 pKa = 3.91QSPFPIIPLIQSKK463 pKa = 9.72SWVPAIVNTIIDD475 pKa = 4.07AEE477 pKa = 4.32EE478 pKa = 4.35KK479 pKa = 10.59GWVPSCSTFKK489 pKa = 10.4IAAIIAMLKK498 pKa = 10.15RR499 pKa = 11.84KK500 pKa = 9.7SSNQGLWNHH509 pKa = 6.51IYY511 pKa = 9.6KK512 pKa = 10.74LSMIHH517 pKa = 6.06SSCYY521 pKa = 10.13LAFKK525 pKa = 10.74DD526 pKa = 4.81KK527 pKa = 10.39ISNMDD532 pKa = 4.92CINMLTDD539 pKa = 3.89AFQIEE544 pKa = 4.42RR545 pKa = 11.84LSQNEE550 pKa = 3.78VSASILIAQASKK562 pKa = 11.25NILATLGKK570 pKa = 10.18EE571 pKa = 3.94IFKK574 pKa = 9.5STLFQLPEE582 pKa = 4.36EE583 pKa = 4.12IQKK586 pKa = 10.32VKK588 pKa = 10.74EE589 pKa = 3.9LVDD592 pKa = 4.15FEE594 pKa = 4.28EE595 pKa = 4.89TTWSKK600 pKa = 11.26YY601 pKa = 10.33SPQEE605 pKa = 3.69HH606 pKa = 6.7EE607 pKa = 3.9MTEE610 pKa = 4.16RR611 pKa = 11.84LPHH614 pKa = 6.57CDD616 pKa = 3.38VLRR619 pKa = 11.84KK620 pKa = 9.47ICGKK624 pKa = 9.83IFEE627 pKa = 5.28PDD629 pKa = 3.07FPGIQDD635 pKa = 3.67EE636 pKa = 4.85PDD638 pKa = 3.48PKK640 pKa = 10.29SWKK643 pKa = 8.33RR644 pKa = 11.84TIRR647 pKa = 11.84GLISFDD653 pKa = 3.44VKK655 pKa = 10.85KK656 pKa = 11.25VFIKK660 pKa = 10.47DD661 pKa = 3.37KK662 pKa = 10.71KK663 pKa = 10.05RR664 pKa = 11.84VEE666 pKa = 3.77ILQVSKK672 pKa = 11.01VSKK675 pKa = 9.84SLCGIIKK682 pKa = 10.15SLNNMEE688 pKa = 4.36EE689 pKa = 5.26KK690 pKa = 10.47EE691 pKa = 4.52LEE693 pKa = 4.11WTNLKK698 pKa = 10.19PGSSSLTSLNYY709 pKa = 10.66LFFPP713 pKa = 5.28
MM1 pKa = 7.7NMNQNKK7 pKa = 9.85AYY9 pKa = 9.89SALVQAIFTFLIYY22 pKa = 9.81YY23 pKa = 10.23SPGFEE28 pKa = 3.92SDD30 pKa = 3.6KK31 pKa = 10.99IVIILSDD38 pKa = 4.0FKK40 pKa = 10.88RR41 pKa = 11.84HH42 pKa = 5.88CDD44 pKa = 3.32QRR46 pKa = 11.84GIKK49 pKa = 10.25AGISWIKK56 pKa = 10.27DD57 pKa = 3.0LRR59 pKa = 11.84LCFTKK64 pKa = 10.93YY65 pKa = 8.91QAGDD69 pKa = 3.59SKK71 pKa = 11.22PKK73 pKa = 10.81VNIKK77 pKa = 9.57CHH79 pKa = 6.27PNGCPRR85 pKa = 11.84LIWDD89 pKa = 3.78NYY91 pKa = 10.41KK92 pKa = 10.88DD93 pKa = 3.72VLNDD97 pKa = 3.52PKK99 pKa = 11.04GIGIIITLLSISRR112 pKa = 11.84PFEE115 pKa = 4.57GIFEE119 pKa = 4.66PSLSDD124 pKa = 3.18ITDD127 pKa = 3.59TPKK130 pKa = 10.55TDD132 pKa = 3.44LGIIKK137 pKa = 10.03DD138 pKa = 3.9LEE140 pKa = 4.17EE141 pKa = 3.9FLPAFFEE148 pKa = 4.61EE149 pKa = 4.95VVHH152 pKa = 5.84VRR154 pKa = 11.84KK155 pKa = 10.07EE156 pKa = 3.89SGQLKK161 pKa = 8.95RR162 pKa = 11.84PKK164 pKa = 9.39WSSWLNNIKK173 pKa = 10.07IAPNGVSINSSYY185 pKa = 11.81ADD187 pKa = 3.48LDD189 pKa = 3.72LHH191 pKa = 7.4NEE193 pKa = 4.04EE194 pKa = 5.2IIRR197 pKa = 11.84DD198 pKa = 3.91LFEE201 pKa = 6.35LGGDD205 pKa = 3.96CFEE208 pKa = 5.42KK209 pKa = 10.34EE210 pKa = 3.91YY211 pKa = 11.46SSIKK215 pKa = 10.47DD216 pKa = 3.29YY217 pKa = 11.04WKK219 pKa = 10.73DD220 pKa = 3.36PEE222 pKa = 4.33GQKK225 pKa = 10.58EE226 pKa = 3.86ITKK229 pKa = 10.31NYY231 pKa = 9.43PIVNTYY237 pKa = 8.83KK238 pKa = 10.41SKK240 pKa = 10.33YY241 pKa = 8.94LGRR244 pKa = 11.84LAVIPSPEE252 pKa = 3.98GKK254 pKa = 9.96SRR256 pKa = 11.84VIAIGDD262 pKa = 3.5YY263 pKa = 9.59WSQNVLFPLHH273 pKa = 6.31NKK275 pKa = 9.41IFGLLKK281 pKa = 10.49RR282 pKa = 11.84IPQDD286 pKa = 3.04LTFAQEE292 pKa = 3.88QGLSKK297 pKa = 10.25IKK299 pKa = 10.12KK300 pKa = 9.97DD301 pKa = 3.43RR302 pKa = 11.84DD303 pKa = 3.19HH304 pKa = 7.15SYY306 pKa = 10.45WSLDD310 pKa = 3.44LKK312 pKa = 11.0SATDD316 pKa = 3.74RR317 pKa = 11.84FPIEE321 pKa = 3.66LQRR324 pKa = 11.84RR325 pKa = 11.84IISYY329 pKa = 9.95IYY331 pKa = 9.91GEE333 pKa = 4.59SYY335 pKa = 10.56SISWRR340 pKa = 11.84NLMSRR345 pKa = 11.84PFYY348 pKa = 9.87QTISGKK354 pKa = 9.86SEE356 pKa = 3.57VTYY359 pKa = 10.61GAGQPIGIYY368 pKa = 10.05SSWSVFTLCHH378 pKa = 6.5HH379 pKa = 6.57FVLYY383 pKa = 10.1CAQRR387 pKa = 11.84RR388 pKa = 11.84SIALKK393 pKa = 9.35KK394 pKa = 9.92GRR396 pKa = 11.84YY397 pKa = 8.78VILGDD402 pKa = 5.28DD403 pKa = 4.11IVISDD408 pKa = 4.43DD409 pKa = 3.6NLAIEE414 pKa = 4.24YY415 pKa = 9.8KK416 pKa = 10.65KK417 pKa = 10.8LINLLGVEE425 pKa = 3.93ISEE428 pKa = 4.5HH429 pKa = 5.4KK430 pKa = 7.73THH432 pKa = 6.93KK433 pKa = 10.26SKK435 pKa = 10.85HH436 pKa = 5.69SYY438 pKa = 10.09EE439 pKa = 3.78IAKK442 pKa = 10.03RR443 pKa = 11.84WITEE447 pKa = 3.86KK448 pKa = 11.04GEE450 pKa = 3.91QSPFPIIPLIQSKK463 pKa = 9.72SWVPAIVNTIIDD475 pKa = 4.07AEE477 pKa = 4.32EE478 pKa = 4.35KK479 pKa = 10.59GWVPSCSTFKK489 pKa = 10.4IAAIIAMLKK498 pKa = 10.15RR499 pKa = 11.84KK500 pKa = 9.7SSNQGLWNHH509 pKa = 6.51IYY511 pKa = 9.6KK512 pKa = 10.74LSMIHH517 pKa = 6.06SSCYY521 pKa = 10.13LAFKK525 pKa = 10.74DD526 pKa = 4.81KK527 pKa = 10.39ISNMDD532 pKa = 4.92CINMLTDD539 pKa = 3.89AFQIEE544 pKa = 4.42RR545 pKa = 11.84LSQNEE550 pKa = 3.78VSASILIAQASKK562 pKa = 11.25NILATLGKK570 pKa = 10.18EE571 pKa = 3.94IFKK574 pKa = 9.5STLFQLPEE582 pKa = 4.36EE583 pKa = 4.12IQKK586 pKa = 10.32VKK588 pKa = 10.74EE589 pKa = 3.9LVDD592 pKa = 4.15FEE594 pKa = 4.28EE595 pKa = 4.89TTWSKK600 pKa = 11.26YY601 pKa = 10.33SPQEE605 pKa = 3.69HH606 pKa = 6.7EE607 pKa = 3.9MTEE610 pKa = 4.16RR611 pKa = 11.84LPHH614 pKa = 6.57CDD616 pKa = 3.38VLRR619 pKa = 11.84KK620 pKa = 9.47ICGKK624 pKa = 9.83IFEE627 pKa = 5.28PDD629 pKa = 3.07FPGIQDD635 pKa = 3.67EE636 pKa = 4.85PDD638 pKa = 3.48PKK640 pKa = 10.29SWKK643 pKa = 8.33RR644 pKa = 11.84TIRR647 pKa = 11.84GLISFDD653 pKa = 3.44VKK655 pKa = 10.85KK656 pKa = 11.25VFIKK660 pKa = 10.47DD661 pKa = 3.37KK662 pKa = 10.71KK663 pKa = 10.05RR664 pKa = 11.84VEE666 pKa = 3.77ILQVSKK672 pKa = 11.01VSKK675 pKa = 9.84SLCGIIKK682 pKa = 10.15SLNNMEE688 pKa = 4.36EE689 pKa = 5.26KK690 pKa = 10.47EE691 pKa = 4.52LEE693 pKa = 4.11WTNLKK698 pKa = 10.19PGSSSLTSLNYY709 pKa = 10.66LFFPP713 pKa = 5.28
Molecular weight: 81.88 kDa
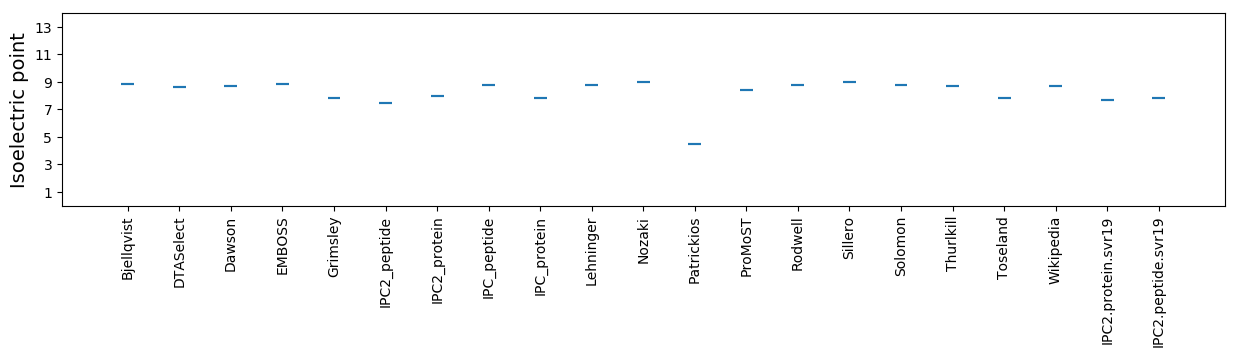
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A191KCP3|A0A191KCP3_9VIRU RNA-dependent RNA polymerase OS=Cronartium ribicola mitovirus 3 OX=1816486 GN=RdRp PE=4 SV=1
MM1 pKa = 7.7NMNQNKK7 pKa = 9.85AYY9 pKa = 9.89SALVQAIFTFLIYY22 pKa = 9.81YY23 pKa = 10.23SPGFEE28 pKa = 3.92SDD30 pKa = 3.6KK31 pKa = 10.99IVIILSDD38 pKa = 4.0FKK40 pKa = 10.88RR41 pKa = 11.84HH42 pKa = 5.88CDD44 pKa = 3.32QRR46 pKa = 11.84GIKK49 pKa = 10.25AGISWIKK56 pKa = 10.27DD57 pKa = 3.0LRR59 pKa = 11.84LCFTKK64 pKa = 10.93YY65 pKa = 8.91QAGDD69 pKa = 3.59SKK71 pKa = 11.22PKK73 pKa = 10.81VNIKK77 pKa = 9.57CHH79 pKa = 6.27PNGCPRR85 pKa = 11.84LIWDD89 pKa = 3.78NYY91 pKa = 10.41KK92 pKa = 10.88DD93 pKa = 3.72VLNDD97 pKa = 3.52PKK99 pKa = 11.04GIGIIITLLSISRR112 pKa = 11.84PFEE115 pKa = 4.57GIFEE119 pKa = 4.66PSLSDD124 pKa = 3.18ITDD127 pKa = 3.59TPKK130 pKa = 10.55TDD132 pKa = 3.44LGIIKK137 pKa = 10.03DD138 pKa = 3.9LEE140 pKa = 4.17EE141 pKa = 3.9FLPAFFEE148 pKa = 4.61EE149 pKa = 4.95VVHH152 pKa = 5.84VRR154 pKa = 11.84KK155 pKa = 10.07EE156 pKa = 3.89SGQLKK161 pKa = 8.95RR162 pKa = 11.84PKK164 pKa = 9.39WSSWLNNIKK173 pKa = 10.07IAPNGVSINSSYY185 pKa = 11.81ADD187 pKa = 3.48LDD189 pKa = 3.72LHH191 pKa = 7.4NEE193 pKa = 4.04EE194 pKa = 5.2IIRR197 pKa = 11.84DD198 pKa = 3.91LFEE201 pKa = 6.35LGGDD205 pKa = 3.96CFEE208 pKa = 5.42KK209 pKa = 10.34EE210 pKa = 3.91YY211 pKa = 11.46SSIKK215 pKa = 10.47DD216 pKa = 3.29YY217 pKa = 11.04WKK219 pKa = 10.73DD220 pKa = 3.36PEE222 pKa = 4.33GQKK225 pKa = 10.58EE226 pKa = 3.86ITKK229 pKa = 10.31NYY231 pKa = 9.43PIVNTYY237 pKa = 8.83KK238 pKa = 10.41SKK240 pKa = 10.33YY241 pKa = 8.94LGRR244 pKa = 11.84LAVIPSPEE252 pKa = 3.98GKK254 pKa = 9.96SRR256 pKa = 11.84VIAIGDD262 pKa = 3.5YY263 pKa = 9.59WSQNVLFPLHH273 pKa = 6.31NKK275 pKa = 9.41IFGLLKK281 pKa = 10.49RR282 pKa = 11.84IPQDD286 pKa = 3.04LTFAQEE292 pKa = 3.88QGLSKK297 pKa = 10.25IKK299 pKa = 10.12KK300 pKa = 9.97DD301 pKa = 3.43RR302 pKa = 11.84DD303 pKa = 3.19HH304 pKa = 7.15SYY306 pKa = 10.45WSLDD310 pKa = 3.44LKK312 pKa = 11.0SATDD316 pKa = 3.74RR317 pKa = 11.84FPIEE321 pKa = 3.66LQRR324 pKa = 11.84RR325 pKa = 11.84IISYY329 pKa = 9.95IYY331 pKa = 9.91GEE333 pKa = 4.59SYY335 pKa = 10.56SISWRR340 pKa = 11.84NLMSRR345 pKa = 11.84PFYY348 pKa = 9.87QTISGKK354 pKa = 9.86SEE356 pKa = 3.57VTYY359 pKa = 10.61GAGQPIGIYY368 pKa = 10.05SSWSVFTLCHH378 pKa = 6.5HH379 pKa = 6.57FVLYY383 pKa = 10.1CAQRR387 pKa = 11.84RR388 pKa = 11.84SIALKK393 pKa = 9.35KK394 pKa = 9.92GRR396 pKa = 11.84YY397 pKa = 8.78VILGDD402 pKa = 5.28DD403 pKa = 4.11IVISDD408 pKa = 4.43DD409 pKa = 3.6NLAIEE414 pKa = 4.24YY415 pKa = 9.8KK416 pKa = 10.65KK417 pKa = 10.8LINLLGVEE425 pKa = 3.93ISEE428 pKa = 4.5HH429 pKa = 5.4KK430 pKa = 7.73THH432 pKa = 6.93KK433 pKa = 10.26SKK435 pKa = 10.85HH436 pKa = 5.69SYY438 pKa = 10.09EE439 pKa = 3.78IAKK442 pKa = 10.03RR443 pKa = 11.84WITEE447 pKa = 3.86KK448 pKa = 11.04GEE450 pKa = 3.91QSPFPIIPLIQSKK463 pKa = 9.72SWVPAIVNTIIDD475 pKa = 4.07AEE477 pKa = 4.32EE478 pKa = 4.35KK479 pKa = 10.59GWVPSCSTFKK489 pKa = 10.4IAAIIAMLKK498 pKa = 10.15RR499 pKa = 11.84KK500 pKa = 9.7SSNQGLWNHH509 pKa = 6.51IYY511 pKa = 9.6KK512 pKa = 10.74LSMIHH517 pKa = 6.06SSCYY521 pKa = 10.13LAFKK525 pKa = 10.74DD526 pKa = 4.81KK527 pKa = 10.39ISNMDD532 pKa = 4.92CINMLTDD539 pKa = 3.89AFQIEE544 pKa = 4.42RR545 pKa = 11.84LSQNEE550 pKa = 3.78VSASILIAQASKK562 pKa = 11.25NILATLGKK570 pKa = 10.18EE571 pKa = 3.94IFKK574 pKa = 9.5STLFQLPEE582 pKa = 4.36EE583 pKa = 4.12IQKK586 pKa = 10.32VKK588 pKa = 10.74EE589 pKa = 3.9LVDD592 pKa = 4.15FEE594 pKa = 4.28EE595 pKa = 4.89TTWSKK600 pKa = 11.26YY601 pKa = 10.33SPQEE605 pKa = 3.69HH606 pKa = 6.7EE607 pKa = 3.9MTEE610 pKa = 4.16RR611 pKa = 11.84LPHH614 pKa = 6.57CDD616 pKa = 3.38VLRR619 pKa = 11.84KK620 pKa = 9.47ICGKK624 pKa = 9.83IFEE627 pKa = 5.28PDD629 pKa = 3.07FPGIQDD635 pKa = 3.67EE636 pKa = 4.85PDD638 pKa = 3.48PKK640 pKa = 10.29SWKK643 pKa = 8.33RR644 pKa = 11.84TIRR647 pKa = 11.84GLISFDD653 pKa = 3.44VKK655 pKa = 10.85KK656 pKa = 11.25VFIKK660 pKa = 10.47DD661 pKa = 3.37KK662 pKa = 10.71KK663 pKa = 10.05RR664 pKa = 11.84VEE666 pKa = 3.77ILQVSKK672 pKa = 11.01VSKK675 pKa = 9.84SLCGIIKK682 pKa = 10.15SLNNMEE688 pKa = 4.36EE689 pKa = 5.26KK690 pKa = 10.47EE691 pKa = 4.52LEE693 pKa = 4.11WTNLKK698 pKa = 10.19PGSSSLTSLNYY709 pKa = 10.66LFFPP713 pKa = 5.28
MM1 pKa = 7.7NMNQNKK7 pKa = 9.85AYY9 pKa = 9.89SALVQAIFTFLIYY22 pKa = 9.81YY23 pKa = 10.23SPGFEE28 pKa = 3.92SDD30 pKa = 3.6KK31 pKa = 10.99IVIILSDD38 pKa = 4.0FKK40 pKa = 10.88RR41 pKa = 11.84HH42 pKa = 5.88CDD44 pKa = 3.32QRR46 pKa = 11.84GIKK49 pKa = 10.25AGISWIKK56 pKa = 10.27DD57 pKa = 3.0LRR59 pKa = 11.84LCFTKK64 pKa = 10.93YY65 pKa = 8.91QAGDD69 pKa = 3.59SKK71 pKa = 11.22PKK73 pKa = 10.81VNIKK77 pKa = 9.57CHH79 pKa = 6.27PNGCPRR85 pKa = 11.84LIWDD89 pKa = 3.78NYY91 pKa = 10.41KK92 pKa = 10.88DD93 pKa = 3.72VLNDD97 pKa = 3.52PKK99 pKa = 11.04GIGIIITLLSISRR112 pKa = 11.84PFEE115 pKa = 4.57GIFEE119 pKa = 4.66PSLSDD124 pKa = 3.18ITDD127 pKa = 3.59TPKK130 pKa = 10.55TDD132 pKa = 3.44LGIIKK137 pKa = 10.03DD138 pKa = 3.9LEE140 pKa = 4.17EE141 pKa = 3.9FLPAFFEE148 pKa = 4.61EE149 pKa = 4.95VVHH152 pKa = 5.84VRR154 pKa = 11.84KK155 pKa = 10.07EE156 pKa = 3.89SGQLKK161 pKa = 8.95RR162 pKa = 11.84PKK164 pKa = 9.39WSSWLNNIKK173 pKa = 10.07IAPNGVSINSSYY185 pKa = 11.81ADD187 pKa = 3.48LDD189 pKa = 3.72LHH191 pKa = 7.4NEE193 pKa = 4.04EE194 pKa = 5.2IIRR197 pKa = 11.84DD198 pKa = 3.91LFEE201 pKa = 6.35LGGDD205 pKa = 3.96CFEE208 pKa = 5.42KK209 pKa = 10.34EE210 pKa = 3.91YY211 pKa = 11.46SSIKK215 pKa = 10.47DD216 pKa = 3.29YY217 pKa = 11.04WKK219 pKa = 10.73DD220 pKa = 3.36PEE222 pKa = 4.33GQKK225 pKa = 10.58EE226 pKa = 3.86ITKK229 pKa = 10.31NYY231 pKa = 9.43PIVNTYY237 pKa = 8.83KK238 pKa = 10.41SKK240 pKa = 10.33YY241 pKa = 8.94LGRR244 pKa = 11.84LAVIPSPEE252 pKa = 3.98GKK254 pKa = 9.96SRR256 pKa = 11.84VIAIGDD262 pKa = 3.5YY263 pKa = 9.59WSQNVLFPLHH273 pKa = 6.31NKK275 pKa = 9.41IFGLLKK281 pKa = 10.49RR282 pKa = 11.84IPQDD286 pKa = 3.04LTFAQEE292 pKa = 3.88QGLSKK297 pKa = 10.25IKK299 pKa = 10.12KK300 pKa = 9.97DD301 pKa = 3.43RR302 pKa = 11.84DD303 pKa = 3.19HH304 pKa = 7.15SYY306 pKa = 10.45WSLDD310 pKa = 3.44LKK312 pKa = 11.0SATDD316 pKa = 3.74RR317 pKa = 11.84FPIEE321 pKa = 3.66LQRR324 pKa = 11.84RR325 pKa = 11.84IISYY329 pKa = 9.95IYY331 pKa = 9.91GEE333 pKa = 4.59SYY335 pKa = 10.56SISWRR340 pKa = 11.84NLMSRR345 pKa = 11.84PFYY348 pKa = 9.87QTISGKK354 pKa = 9.86SEE356 pKa = 3.57VTYY359 pKa = 10.61GAGQPIGIYY368 pKa = 10.05SSWSVFTLCHH378 pKa = 6.5HH379 pKa = 6.57FVLYY383 pKa = 10.1CAQRR387 pKa = 11.84RR388 pKa = 11.84SIALKK393 pKa = 9.35KK394 pKa = 9.92GRR396 pKa = 11.84YY397 pKa = 8.78VILGDD402 pKa = 5.28DD403 pKa = 4.11IVISDD408 pKa = 4.43DD409 pKa = 3.6NLAIEE414 pKa = 4.24YY415 pKa = 9.8KK416 pKa = 10.65KK417 pKa = 10.8LINLLGVEE425 pKa = 3.93ISEE428 pKa = 4.5HH429 pKa = 5.4KK430 pKa = 7.73THH432 pKa = 6.93KK433 pKa = 10.26SKK435 pKa = 10.85HH436 pKa = 5.69SYY438 pKa = 10.09EE439 pKa = 3.78IAKK442 pKa = 10.03RR443 pKa = 11.84WITEE447 pKa = 3.86KK448 pKa = 11.04GEE450 pKa = 3.91QSPFPIIPLIQSKK463 pKa = 9.72SWVPAIVNTIIDD475 pKa = 4.07AEE477 pKa = 4.32EE478 pKa = 4.35KK479 pKa = 10.59GWVPSCSTFKK489 pKa = 10.4IAAIIAMLKK498 pKa = 10.15RR499 pKa = 11.84KK500 pKa = 9.7SSNQGLWNHH509 pKa = 6.51IYY511 pKa = 9.6KK512 pKa = 10.74LSMIHH517 pKa = 6.06SSCYY521 pKa = 10.13LAFKK525 pKa = 10.74DD526 pKa = 4.81KK527 pKa = 10.39ISNMDD532 pKa = 4.92CINMLTDD539 pKa = 3.89AFQIEE544 pKa = 4.42RR545 pKa = 11.84LSQNEE550 pKa = 3.78VSASILIAQASKK562 pKa = 11.25NILATLGKK570 pKa = 10.18EE571 pKa = 3.94IFKK574 pKa = 9.5STLFQLPEE582 pKa = 4.36EE583 pKa = 4.12IQKK586 pKa = 10.32VKK588 pKa = 10.74EE589 pKa = 3.9LVDD592 pKa = 4.15FEE594 pKa = 4.28EE595 pKa = 4.89TTWSKK600 pKa = 11.26YY601 pKa = 10.33SPQEE605 pKa = 3.69HH606 pKa = 6.7EE607 pKa = 3.9MTEE610 pKa = 4.16RR611 pKa = 11.84LPHH614 pKa = 6.57CDD616 pKa = 3.38VLRR619 pKa = 11.84KK620 pKa = 9.47ICGKK624 pKa = 9.83IFEE627 pKa = 5.28PDD629 pKa = 3.07FPGIQDD635 pKa = 3.67EE636 pKa = 4.85PDD638 pKa = 3.48PKK640 pKa = 10.29SWKK643 pKa = 8.33RR644 pKa = 11.84TIRR647 pKa = 11.84GLISFDD653 pKa = 3.44VKK655 pKa = 10.85KK656 pKa = 11.25VFIKK660 pKa = 10.47DD661 pKa = 3.37KK662 pKa = 10.71KK663 pKa = 10.05RR664 pKa = 11.84VEE666 pKa = 3.77ILQVSKK672 pKa = 11.01VSKK675 pKa = 9.84SLCGIIKK682 pKa = 10.15SLNNMEE688 pKa = 4.36EE689 pKa = 5.26KK690 pKa = 10.47EE691 pKa = 4.52LEE693 pKa = 4.11WTNLKK698 pKa = 10.19PGSSSLTSLNYY709 pKa = 10.66LFFPP713 pKa = 5.28
Molecular weight: 81.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
713 |
713 |
713 |
713.0 |
81.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.927 ± 0.0 | 1.823 ± 0.0 |
5.47 ± 0.0 | 6.452 ± 0.0 |
4.488 ± 0.0 | 5.049 ± 0.0 |
2.104 ± 0.0 | 10.94 ± 0.0 |
9.677 ± 0.0 | 8.836 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.262 ± 0.0 | 4.208 ± 0.0 |
4.769 ± 0.0 | 3.506 ± 0.0 |
3.927 ± 0.0 | 9.677 ± 0.0 |
3.647 ± 0.0 | 4.208 ± 0.0 |
2.244 ± 0.0 | 3.787 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
