
Leviviridae sp.
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Allassoviricetes; Levivirales; Leviviridae; unclassified Leviviridae
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
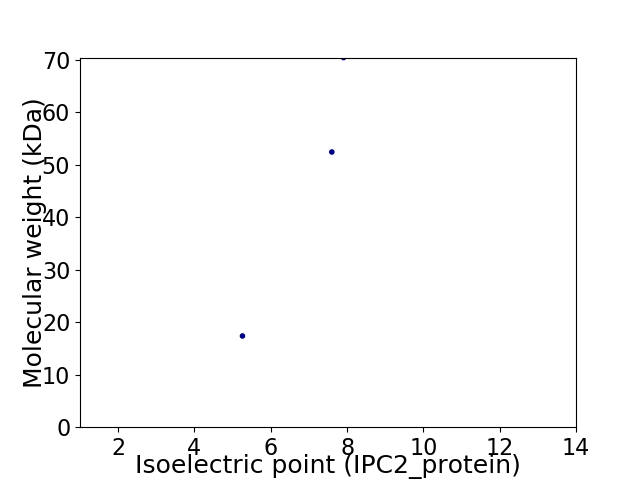
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
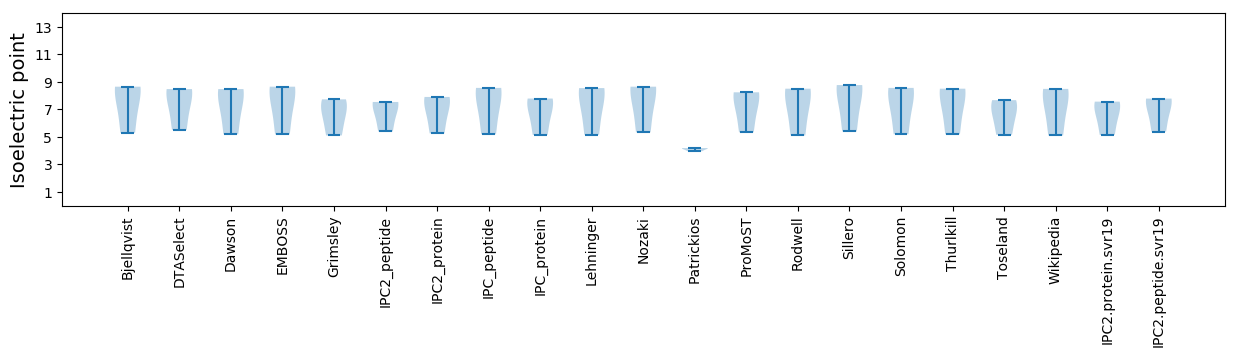
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A142D861|A0A142D861_9VIRU RNA replicase beta chain OS=Leviviridae sp. OX=2027243 PE=4 SV=1
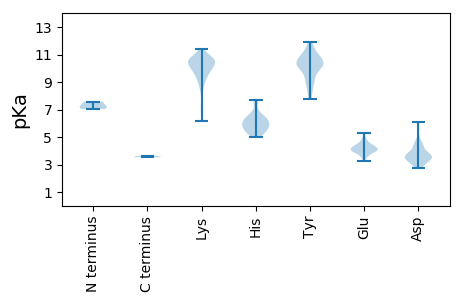
MM1 pKa = 7.58SKK3 pKa = 9.59TSVIDD8 pKa = 3.46ATNTTASTTFNASAYY23 pKa = 8.94KK24 pKa = 10.57LSVLGNYY31 pKa = 10.18ALTADD36 pKa = 4.45EE37 pKa = 4.54PTEE40 pKa = 3.88ATYY43 pKa = 11.63SNVTSSIEE51 pKa = 4.0RR52 pKa = 11.84PEE54 pKa = 3.85LLQYY58 pKa = 10.54RR59 pKa = 11.84SRR61 pKa = 11.84RR62 pKa = 11.84SEE64 pKa = 3.93KK65 pKa = 10.88NNTGLNVPNVSDD77 pKa = 3.25TAGYY81 pKa = 9.34VEE83 pKa = 4.52YY84 pKa = 10.54GIRR87 pKa = 11.84LQNVVHH93 pKa = 5.72TTDD96 pKa = 3.06TADD99 pKa = 3.62ANVEE103 pKa = 4.03IADD106 pKa = 4.05PCCVNITLKK115 pKa = 10.89HH116 pKa = 5.86PATSRR121 pKa = 11.84ITEE124 pKa = 4.76DD125 pKa = 2.86IATQLLMRR133 pKa = 11.84AVSTMFDD140 pKa = 3.41DD141 pKa = 4.33AGKK144 pKa = 10.63SRR146 pKa = 11.84LGDD149 pKa = 3.32MMRR152 pKa = 11.84SALHH156 pKa = 5.18ITANN160 pKa = 3.56
MM1 pKa = 7.58SKK3 pKa = 9.59TSVIDD8 pKa = 3.46ATNTTASTTFNASAYY23 pKa = 8.94KK24 pKa = 10.57LSVLGNYY31 pKa = 10.18ALTADD36 pKa = 4.45EE37 pKa = 4.54PTEE40 pKa = 3.88ATYY43 pKa = 11.63SNVTSSIEE51 pKa = 4.0RR52 pKa = 11.84PEE54 pKa = 3.85LLQYY58 pKa = 10.54RR59 pKa = 11.84SRR61 pKa = 11.84RR62 pKa = 11.84SEE64 pKa = 3.93KK65 pKa = 10.88NNTGLNVPNVSDD77 pKa = 3.25TAGYY81 pKa = 9.34VEE83 pKa = 4.52YY84 pKa = 10.54GIRR87 pKa = 11.84LQNVVHH93 pKa = 5.72TTDD96 pKa = 3.06TADD99 pKa = 3.62ANVEE103 pKa = 4.03IADD106 pKa = 4.05PCCVNITLKK115 pKa = 10.89HH116 pKa = 5.86PATSRR121 pKa = 11.84ITEE124 pKa = 4.76DD125 pKa = 2.86IATQLLMRR133 pKa = 11.84AVSTMFDD140 pKa = 3.41DD141 pKa = 4.33AGKK144 pKa = 10.63SRR146 pKa = 11.84LGDD149 pKa = 3.32MMRR152 pKa = 11.84SALHH156 pKa = 5.18ITANN160 pKa = 3.56
Molecular weight: 17.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142D861|A0A142D861_9VIRU RNA replicase beta chain OS=Leviviridae sp. OX=2027243 PE=4 SV=1
MM1 pKa = 7.22YY2 pKa = 10.62LNRR5 pKa = 11.84DD6 pKa = 3.57TVKK9 pKa = 10.78KK10 pKa = 10.33ILALDD15 pKa = 3.74NVEE18 pKa = 4.08VTEE21 pKa = 5.29DD22 pKa = 3.7NIFSAKK28 pKa = 9.87LALEE32 pKa = 4.52VWQSLLSEE40 pKa = 4.3QNPQYY45 pKa = 11.15GKK47 pKa = 10.82LVLSYY52 pKa = 11.06ALRR55 pKa = 11.84YY56 pKa = 8.98GLRR59 pKa = 11.84RR60 pKa = 11.84TIQEE64 pKa = 3.91ACDD67 pKa = 3.53CSEE70 pKa = 3.8ILMGRR75 pKa = 11.84SFRR78 pKa = 11.84NNAFTGIVATATDD91 pKa = 3.94EE92 pKa = 4.8RR93 pKa = 11.84SVLQCLRR100 pKa = 11.84FLKK103 pKa = 10.31RR104 pKa = 11.84FCYY107 pKa = 10.16PDD109 pKa = 4.95DD110 pKa = 5.25DD111 pKa = 4.39SLRR114 pKa = 11.84EE115 pKa = 3.88EE116 pKa = 4.42AVSQFFKK123 pKa = 10.14RR124 pKa = 11.84THH126 pKa = 5.0GWAQLKK132 pKa = 10.44ISDD135 pKa = 4.16YY136 pKa = 10.02LRR138 pKa = 11.84SVIKK142 pKa = 10.71SKK144 pKa = 10.83FSDD147 pKa = 3.01IVGSEE152 pKa = 3.57KK153 pKa = 10.41NYY155 pKa = 10.15RR156 pKa = 11.84RR157 pKa = 11.84SEE159 pKa = 4.08VANLTKK165 pKa = 10.35SYY167 pKa = 11.17FDD169 pKa = 3.79IPSGAAYY176 pKa = 10.16DD177 pKa = 3.65IEE179 pKa = 4.35YY180 pKa = 10.83DD181 pKa = 3.59RR182 pKa = 11.84LVRR185 pKa = 11.84TQGEE189 pKa = 4.64KK190 pKa = 9.02ITILNEE196 pKa = 3.67YY197 pKa = 9.01VKK199 pKa = 10.0YY200 pKa = 10.19HH201 pKa = 6.55PSYY204 pKa = 10.37PGIKK208 pKa = 9.52RR209 pKa = 11.84FVYY212 pKa = 10.23DD213 pKa = 3.16QSKK216 pKa = 10.95DD217 pKa = 3.34FTPISKK223 pKa = 10.68LIAVPKK229 pKa = 9.69NYY231 pKa = 7.92KK232 pKa = 7.08TSRR235 pKa = 11.84VIAVEE240 pKa = 4.01HH241 pKa = 6.63PLVQFRR247 pKa = 11.84LGAISNYY254 pKa = 10.18LSDD257 pKa = 6.07RR258 pKa = 11.84IMKK261 pKa = 7.21YY262 pKa = 8.5TKK264 pKa = 10.47GRR266 pKa = 11.84INIQDD271 pKa = 3.13QSRR274 pKa = 11.84NRR276 pKa = 11.84NLAQLGSAQDD286 pKa = 3.48SYY288 pKa = 11.95ATIDD292 pKa = 3.81LSAASDD298 pKa = 3.98SNTQMLQYY306 pKa = 9.59QICPDD311 pKa = 2.91WLYY314 pKa = 11.33RR315 pKa = 11.84EE316 pKa = 3.97ISPWISRR323 pKa = 11.84RR324 pKa = 11.84VRR326 pKa = 11.84YY327 pKa = 9.4NGKK330 pKa = 6.2TRR332 pKa = 11.84RR333 pKa = 11.84VNTFGTMGTRR343 pKa = 11.84ITFPLEE349 pKa = 3.69MVTFIAVVEE358 pKa = 4.31VARR361 pKa = 11.84DD362 pKa = 3.72YY363 pKa = 11.74YY364 pKa = 9.68NTLVGEE370 pKa = 3.96PHH372 pKa = 6.32ITLDD376 pKa = 4.6DD377 pKa = 3.58IAVYY381 pKa = 10.59GDD383 pKa = 4.61DD384 pKa = 3.76IIVPKK389 pKa = 10.29EE390 pKa = 3.84LVSTVSDD397 pKa = 3.63LLGILGFTVNAEE409 pKa = 4.15KK410 pKa = 9.89TYY412 pKa = 10.51SSGPFRR418 pKa = 11.84EE419 pKa = 4.46SCGGDD424 pKa = 3.45YY425 pKa = 11.29YY426 pKa = 11.64NGVDD430 pKa = 3.57VSTNYY435 pKa = 9.4WPRR438 pKa = 11.84TPIRR442 pKa = 11.84RR443 pKa = 11.84DD444 pKa = 2.74ISTIPSLISLQHH456 pKa = 6.4RR457 pKa = 11.84LVQHH461 pKa = 6.23FEE463 pKa = 4.39SNLILVNTIRR473 pKa = 11.84DD474 pKa = 3.7IVPSITEE481 pKa = 4.02SEE483 pKa = 3.97IGSPYY488 pKa = 8.91TDD490 pKa = 3.39IWSAWPVLTLRR501 pKa = 11.84EE502 pKa = 4.12GRR504 pKa = 11.84EE505 pKa = 3.39RR506 pKa = 11.84RR507 pKa = 11.84YY508 pKa = 10.87ALTYY512 pKa = 10.99SKK514 pKa = 10.49FSDD517 pKa = 3.51AMRR520 pKa = 11.84PDD522 pKa = 3.24GKK524 pKa = 10.52RR525 pKa = 11.84YY526 pKa = 8.74VAVRR530 pKa = 11.84RR531 pKa = 11.84TKK533 pKa = 10.31KK534 pKa = 9.01IHH536 pKa = 5.97HH537 pKa = 6.21VQTATAEE544 pKa = 4.19YY545 pKa = 10.22HH546 pKa = 5.6STFPPKK552 pKa = 9.48WKK554 pKa = 10.47NGWPSADD561 pKa = 2.94IQEE564 pKa = 4.25FLMYY568 pKa = 10.29RR569 pKa = 11.84SLYY572 pKa = 10.62HH573 pKa = 6.1GGCYY577 pKa = 10.2ASGLDD582 pKa = 3.85EE583 pKa = 4.93LLHH586 pKa = 5.53VTDD589 pKa = 5.69KK590 pKa = 10.82PLHH593 pKa = 5.48QEE595 pKa = 3.24WDD597 pKa = 3.76LPILDD602 pKa = 4.25RR603 pKa = 11.84DD604 pKa = 3.8HH605 pKa = 7.48VRR607 pKa = 11.84VLIEE611 pKa = 3.63
MM1 pKa = 7.22YY2 pKa = 10.62LNRR5 pKa = 11.84DD6 pKa = 3.57TVKK9 pKa = 10.78KK10 pKa = 10.33ILALDD15 pKa = 3.74NVEE18 pKa = 4.08VTEE21 pKa = 5.29DD22 pKa = 3.7NIFSAKK28 pKa = 9.87LALEE32 pKa = 4.52VWQSLLSEE40 pKa = 4.3QNPQYY45 pKa = 11.15GKK47 pKa = 10.82LVLSYY52 pKa = 11.06ALRR55 pKa = 11.84YY56 pKa = 8.98GLRR59 pKa = 11.84RR60 pKa = 11.84TIQEE64 pKa = 3.91ACDD67 pKa = 3.53CSEE70 pKa = 3.8ILMGRR75 pKa = 11.84SFRR78 pKa = 11.84NNAFTGIVATATDD91 pKa = 3.94EE92 pKa = 4.8RR93 pKa = 11.84SVLQCLRR100 pKa = 11.84FLKK103 pKa = 10.31RR104 pKa = 11.84FCYY107 pKa = 10.16PDD109 pKa = 4.95DD110 pKa = 5.25DD111 pKa = 4.39SLRR114 pKa = 11.84EE115 pKa = 3.88EE116 pKa = 4.42AVSQFFKK123 pKa = 10.14RR124 pKa = 11.84THH126 pKa = 5.0GWAQLKK132 pKa = 10.44ISDD135 pKa = 4.16YY136 pKa = 10.02LRR138 pKa = 11.84SVIKK142 pKa = 10.71SKK144 pKa = 10.83FSDD147 pKa = 3.01IVGSEE152 pKa = 3.57KK153 pKa = 10.41NYY155 pKa = 10.15RR156 pKa = 11.84RR157 pKa = 11.84SEE159 pKa = 4.08VANLTKK165 pKa = 10.35SYY167 pKa = 11.17FDD169 pKa = 3.79IPSGAAYY176 pKa = 10.16DD177 pKa = 3.65IEE179 pKa = 4.35YY180 pKa = 10.83DD181 pKa = 3.59RR182 pKa = 11.84LVRR185 pKa = 11.84TQGEE189 pKa = 4.64KK190 pKa = 9.02ITILNEE196 pKa = 3.67YY197 pKa = 9.01VKK199 pKa = 10.0YY200 pKa = 10.19HH201 pKa = 6.55PSYY204 pKa = 10.37PGIKK208 pKa = 9.52RR209 pKa = 11.84FVYY212 pKa = 10.23DD213 pKa = 3.16QSKK216 pKa = 10.95DD217 pKa = 3.34FTPISKK223 pKa = 10.68LIAVPKK229 pKa = 9.69NYY231 pKa = 7.92KK232 pKa = 7.08TSRR235 pKa = 11.84VIAVEE240 pKa = 4.01HH241 pKa = 6.63PLVQFRR247 pKa = 11.84LGAISNYY254 pKa = 10.18LSDD257 pKa = 6.07RR258 pKa = 11.84IMKK261 pKa = 7.21YY262 pKa = 8.5TKK264 pKa = 10.47GRR266 pKa = 11.84INIQDD271 pKa = 3.13QSRR274 pKa = 11.84NRR276 pKa = 11.84NLAQLGSAQDD286 pKa = 3.48SYY288 pKa = 11.95ATIDD292 pKa = 3.81LSAASDD298 pKa = 3.98SNTQMLQYY306 pKa = 9.59QICPDD311 pKa = 2.91WLYY314 pKa = 11.33RR315 pKa = 11.84EE316 pKa = 3.97ISPWISRR323 pKa = 11.84RR324 pKa = 11.84VRR326 pKa = 11.84YY327 pKa = 9.4NGKK330 pKa = 6.2TRR332 pKa = 11.84RR333 pKa = 11.84VNTFGTMGTRR343 pKa = 11.84ITFPLEE349 pKa = 3.69MVTFIAVVEE358 pKa = 4.31VARR361 pKa = 11.84DD362 pKa = 3.72YY363 pKa = 11.74YY364 pKa = 9.68NTLVGEE370 pKa = 3.96PHH372 pKa = 6.32ITLDD376 pKa = 4.6DD377 pKa = 3.58IAVYY381 pKa = 10.59GDD383 pKa = 4.61DD384 pKa = 3.76IIVPKK389 pKa = 10.29EE390 pKa = 3.84LVSTVSDD397 pKa = 3.63LLGILGFTVNAEE409 pKa = 4.15KK410 pKa = 9.89TYY412 pKa = 10.51SSGPFRR418 pKa = 11.84EE419 pKa = 4.46SCGGDD424 pKa = 3.45YY425 pKa = 11.29YY426 pKa = 11.64NGVDD430 pKa = 3.57VSTNYY435 pKa = 9.4WPRR438 pKa = 11.84TPIRR442 pKa = 11.84RR443 pKa = 11.84DD444 pKa = 2.74ISTIPSLISLQHH456 pKa = 6.4RR457 pKa = 11.84LVQHH461 pKa = 6.23FEE463 pKa = 4.39SNLILVNTIRR473 pKa = 11.84DD474 pKa = 3.7IVPSITEE481 pKa = 4.02SEE483 pKa = 3.97IGSPYY488 pKa = 8.91TDD490 pKa = 3.39IWSAWPVLTLRR501 pKa = 11.84EE502 pKa = 4.12GRR504 pKa = 11.84EE505 pKa = 3.39RR506 pKa = 11.84RR507 pKa = 11.84YY508 pKa = 10.87ALTYY512 pKa = 10.99SKK514 pKa = 10.49FSDD517 pKa = 3.51AMRR520 pKa = 11.84PDD522 pKa = 3.24GKK524 pKa = 10.52RR525 pKa = 11.84YY526 pKa = 8.74VAVRR530 pKa = 11.84RR531 pKa = 11.84TKK533 pKa = 10.31KK534 pKa = 9.01IHH536 pKa = 5.97HH537 pKa = 6.21VQTATAEE544 pKa = 4.19YY545 pKa = 10.22HH546 pKa = 5.6STFPPKK552 pKa = 9.48WKK554 pKa = 10.47NGWPSADD561 pKa = 2.94IQEE564 pKa = 4.25FLMYY568 pKa = 10.29RR569 pKa = 11.84SLYY572 pKa = 10.62HH573 pKa = 6.1GGCYY577 pKa = 10.2ASGLDD582 pKa = 3.85EE583 pKa = 4.93LLHH586 pKa = 5.53VTDD589 pKa = 5.69KK590 pKa = 10.82PLHH593 pKa = 5.48QEE595 pKa = 3.24WDD597 pKa = 3.76LPILDD602 pKa = 4.25RR603 pKa = 11.84DD604 pKa = 3.8HH605 pKa = 7.48VRR607 pKa = 11.84VLIEE611 pKa = 3.63
Molecular weight: 70.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1230 |
160 |
611 |
410.0 |
46.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.26 ± 1.052 | 1.545 ± 0.324 |
6.098 ± 0.333 | 5.854 ± 0.58 |
3.74 ± 0.814 | 5.528 ± 0.796 |
1.951 ± 0.113 | 5.772 ± 1.012 |
5.285 ± 0.76 | 8.537 ± 0.258 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.545 ± 0.385 | 4.228 ± 0.824 |
3.415 ± 0.546 | 3.252 ± 0.336 |
7.236 ± 0.44 | 8.374 ± 0.269 |
7.642 ± 1.326 | 6.911 ± 0.031 |
1.382 ± 0.333 | 5.447 ± 0.416 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
