
Mangrovimonas yunxiaonensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Mangrovimonas
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
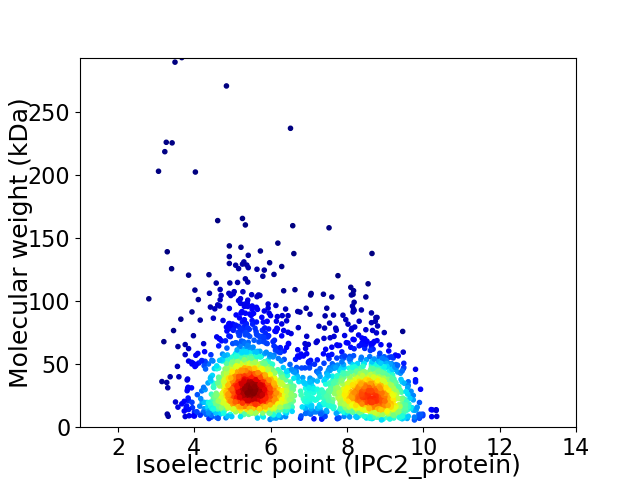
Virtual 2D-PAGE plot for 2307 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
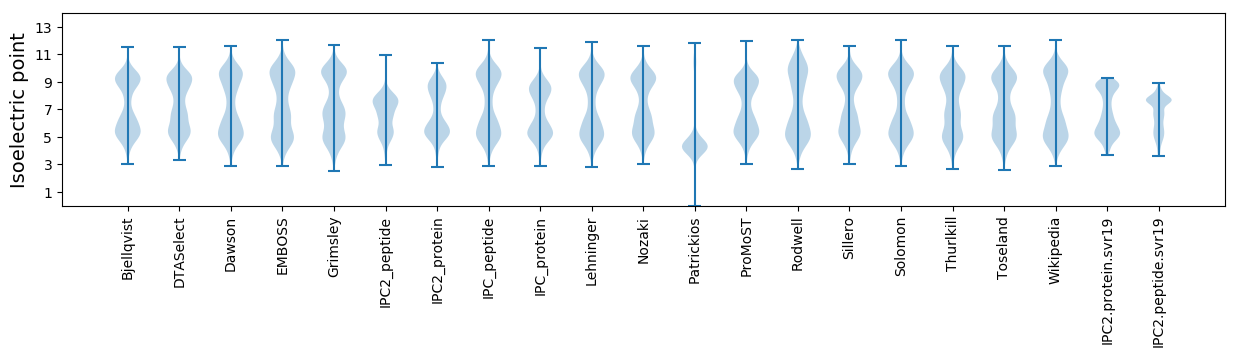
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A084TJU7|A0A084TJU7_9FLAO Uncharacterized protein OS=Mangrovimonas yunxiaonensis OX=1197477 GN=IA57_11140 PE=4 SV=1
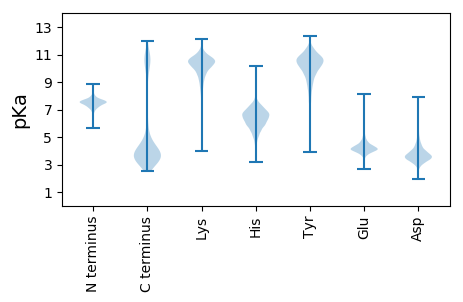
MM1 pKa = 7.49KK2 pKa = 10.3NSNYY6 pKa = 9.9SRR8 pKa = 11.84LVSIFITLLTLVTSCSSDD26 pKa = 4.08DD27 pKa = 4.74DD28 pKa = 4.06NTSNDD33 pKa = 3.89PNNSSTFLDD42 pKa = 3.51KK43 pKa = 10.0WWYY46 pKa = 10.56DD47 pKa = 3.31SDD49 pKa = 5.55DD50 pKa = 3.86FAADD54 pKa = 3.39IYY56 pKa = 10.68FHH58 pKa = 7.37SNGEE62 pKa = 4.09YY63 pKa = 8.51EE64 pKa = 4.23QKK66 pKa = 10.7KK67 pKa = 8.73VVQGTSYY74 pKa = 10.3IANGDD79 pKa = 3.15WVYY82 pKa = 11.07EE83 pKa = 4.13NEE85 pKa = 4.03SSGIIRR91 pKa = 11.84IDD93 pKa = 3.38NLVGNGQTQSTIWLKK108 pKa = 10.85VSNIQVNTITIQQSSDD124 pKa = 2.92GTDD127 pKa = 3.14YY128 pKa = 11.53SEE130 pKa = 5.44AMDD133 pKa = 4.75YY134 pKa = 11.17QDD136 pKa = 3.81TDD138 pKa = 3.08NN139 pKa = 4.62
MM1 pKa = 7.49KK2 pKa = 10.3NSNYY6 pKa = 9.9SRR8 pKa = 11.84LVSIFITLLTLVTSCSSDD26 pKa = 4.08DD27 pKa = 4.74DD28 pKa = 4.06NTSNDD33 pKa = 3.89PNNSSTFLDD42 pKa = 3.51KK43 pKa = 10.0WWYY46 pKa = 10.56DD47 pKa = 3.31SDD49 pKa = 5.55DD50 pKa = 3.86FAADD54 pKa = 3.39IYY56 pKa = 10.68FHH58 pKa = 7.37SNGEE62 pKa = 4.09YY63 pKa = 8.51EE64 pKa = 4.23QKK66 pKa = 10.7KK67 pKa = 8.73VVQGTSYY74 pKa = 10.3IANGDD79 pKa = 3.15WVYY82 pKa = 11.07EE83 pKa = 4.13NEE85 pKa = 4.03SSGIIRR91 pKa = 11.84IDD93 pKa = 3.38NLVGNGQTQSTIWLKK108 pKa = 10.85VSNIQVNTITIQQSSDD124 pKa = 2.92GTDD127 pKa = 3.14YY128 pKa = 11.53SEE130 pKa = 5.44AMDD133 pKa = 4.75YY134 pKa = 11.17QDD136 pKa = 3.81TDD138 pKa = 3.08NN139 pKa = 4.62
Molecular weight: 15.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A084TID8|A0A084TID8_9FLAO 50S ribosomal protein L29 OS=Mangrovimonas yunxiaonensis OX=1197477 GN=rpmC PE=3 SV=1
MM1 pKa = 7.32SVKK4 pKa = 9.96KK5 pKa = 10.34LKK7 pKa = 10.43PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGYY21 pKa = 9.53DD22 pKa = 4.32AITTDD27 pKa = 3.72KK28 pKa = 11.01PEE30 pKa = 4.39KK31 pKa = 10.55SLLAPKK37 pKa = 9.91KK38 pKa = 10.22RR39 pKa = 11.84SGGRR43 pKa = 11.84NSQGKK48 pKa = 6.48MTMRR52 pKa = 11.84YY53 pKa = 9.3IGGGHH58 pKa = 6.61KK59 pKa = 9.72KK60 pKa = 9.77RR61 pKa = 11.84YY62 pKa = 9.44RR63 pKa = 11.84IIDD66 pKa = 3.78FKK68 pKa = 11.1RR69 pKa = 11.84NKK71 pKa = 9.94AGVSAEE77 pKa = 4.05VMSIEE82 pKa = 3.98YY83 pKa = 10.47DD84 pKa = 3.48PNRR87 pKa = 11.84TAFIALLKK95 pKa = 10.95YY96 pKa = 10.31EE97 pKa = 4.98DD98 pKa = 3.76GEE100 pKa = 4.02KK101 pKa = 10.3RR102 pKa = 11.84YY103 pKa = 10.22IIAQNGLQVGQNVISGQEE121 pKa = 3.89NVPTEE126 pKa = 4.12IGNTMPLSKK135 pKa = 10.29IPLGTIISCVEE146 pKa = 3.63LRR148 pKa = 11.84PGQGAVMARR157 pKa = 11.84SAGAFAQLMARR168 pKa = 11.84DD169 pKa = 4.26GKK171 pKa = 10.62FATIKK176 pKa = 10.5LPSGEE181 pKa = 4.11TRR183 pKa = 11.84LVLVSCMATIGVVSNSDD200 pKa = 3.21HH201 pKa = 6.09QLLVSGKK208 pKa = 9.49AGRR211 pKa = 11.84SRR213 pKa = 11.84WLGRR217 pKa = 11.84RR218 pKa = 11.84PRR220 pKa = 11.84TRR222 pKa = 11.84PVVMNPVDD230 pKa = 3.47HH231 pKa = 7.05PMGGGEE237 pKa = 3.89GRR239 pKa = 11.84ASGGHH244 pKa = 5.06PRR246 pKa = 11.84SRR248 pKa = 11.84KK249 pKa = 8.85GLPAKK254 pKa = 10.34GYY256 pKa = 7.1RR257 pKa = 11.84TRR259 pKa = 11.84SKK261 pKa = 9.56TKK263 pKa = 10.12ASNKK267 pKa = 9.84YY268 pKa = 8.91IVEE271 pKa = 4.08RR272 pKa = 11.84RR273 pKa = 11.84KK274 pKa = 10.24KK275 pKa = 10.1
MM1 pKa = 7.32SVKK4 pKa = 9.96KK5 pKa = 10.34LKK7 pKa = 10.43PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGYY21 pKa = 9.53DD22 pKa = 4.32AITTDD27 pKa = 3.72KK28 pKa = 11.01PEE30 pKa = 4.39KK31 pKa = 10.55SLLAPKK37 pKa = 9.91KK38 pKa = 10.22RR39 pKa = 11.84SGGRR43 pKa = 11.84NSQGKK48 pKa = 6.48MTMRR52 pKa = 11.84YY53 pKa = 9.3IGGGHH58 pKa = 6.61KK59 pKa = 9.72KK60 pKa = 9.77RR61 pKa = 11.84YY62 pKa = 9.44RR63 pKa = 11.84IIDD66 pKa = 3.78FKK68 pKa = 11.1RR69 pKa = 11.84NKK71 pKa = 9.94AGVSAEE77 pKa = 4.05VMSIEE82 pKa = 3.98YY83 pKa = 10.47DD84 pKa = 3.48PNRR87 pKa = 11.84TAFIALLKK95 pKa = 10.95YY96 pKa = 10.31EE97 pKa = 4.98DD98 pKa = 3.76GEE100 pKa = 4.02KK101 pKa = 10.3RR102 pKa = 11.84YY103 pKa = 10.22IIAQNGLQVGQNVISGQEE121 pKa = 3.89NVPTEE126 pKa = 4.12IGNTMPLSKK135 pKa = 10.29IPLGTIISCVEE146 pKa = 3.63LRR148 pKa = 11.84PGQGAVMARR157 pKa = 11.84SAGAFAQLMARR168 pKa = 11.84DD169 pKa = 4.26GKK171 pKa = 10.62FATIKK176 pKa = 10.5LPSGEE181 pKa = 4.11TRR183 pKa = 11.84LVLVSCMATIGVVSNSDD200 pKa = 3.21HH201 pKa = 6.09QLLVSGKK208 pKa = 9.49AGRR211 pKa = 11.84SRR213 pKa = 11.84WLGRR217 pKa = 11.84RR218 pKa = 11.84PRR220 pKa = 11.84TRR222 pKa = 11.84PVVMNPVDD230 pKa = 3.47HH231 pKa = 7.05PMGGGEE237 pKa = 3.89GRR239 pKa = 11.84ASGGHH244 pKa = 5.06PRR246 pKa = 11.84SRR248 pKa = 11.84KK249 pKa = 8.85GLPAKK254 pKa = 10.34GYY256 pKa = 7.1RR257 pKa = 11.84TRR259 pKa = 11.84SKK261 pKa = 9.56TKK263 pKa = 10.12ASNKK267 pKa = 9.84YY268 pKa = 8.91IVEE271 pKa = 4.08RR272 pKa = 11.84RR273 pKa = 11.84KK274 pKa = 10.24KK275 pKa = 10.1
Molecular weight: 30.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
774474 |
47 |
2785 |
335.7 |
37.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.16 ± 0.052 | 0.801 ± 0.025 |
5.531 ± 0.056 | 6.269 ± 0.055 |
5.071 ± 0.043 | 6.323 ± 0.054 |
2.041 ± 0.028 | 7.391 ± 0.048 |
7.577 ± 0.076 | 9.449 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.213 ± 0.028 | 6.055 ± 0.053 |
3.473 ± 0.03 | 3.652 ± 0.031 |
3.313 ± 0.039 | 5.922 ± 0.047 |
6.2 ± 0.065 | 6.566 ± 0.04 |
0.969 ± 0.018 | 4.026 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
