
Pigeonpox virus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Pokkesviricetes; Chitovirales; Poxviridae; Chordopoxvirinae; Avipoxvirus
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
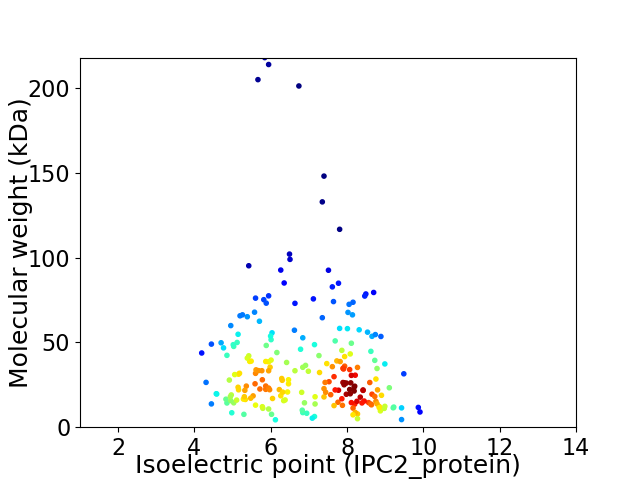
Virtual 2D-PAGE plot for 222 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
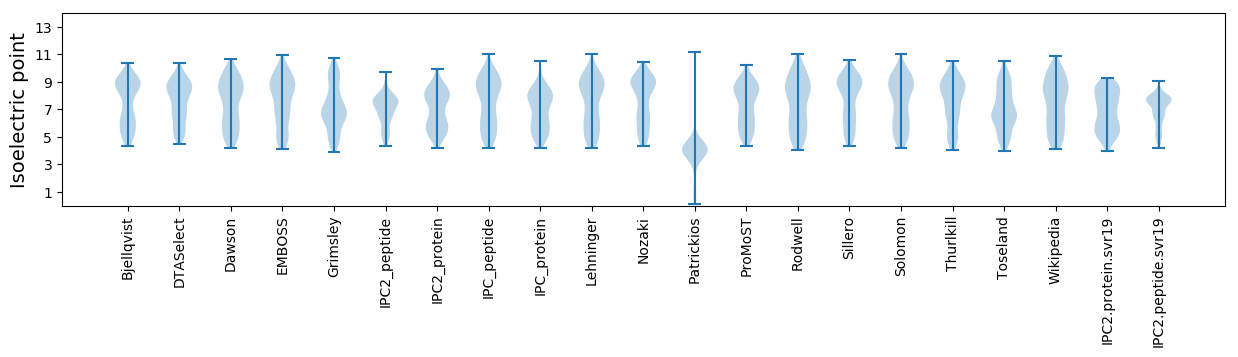
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A068EER1|A0A068EER1_9POXV Major core protein 4a precursor OS=Pigeonpox virus OX=10264 GN=fep_178 PE=3 SV=1
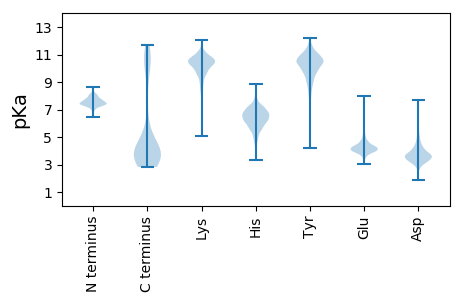
MM1 pKa = 7.76IIYY4 pKa = 10.24IITLSLLFIKK14 pKa = 10.68VITSSKK20 pKa = 10.48EE21 pKa = 3.87PVKK24 pKa = 10.83NPQDD28 pKa = 3.13ILHH31 pKa = 6.73IIEE34 pKa = 4.42HH35 pKa = 6.23NKK37 pKa = 9.44TGVTACSLYY46 pKa = 11.02CFDD49 pKa = 4.71SSKK52 pKa = 11.34GLDD55 pKa = 3.42QPKK58 pKa = 8.87TFILPGKK65 pKa = 9.66YY66 pKa = 9.99SNNSIKK72 pKa = 10.87LEE74 pKa = 4.03VAIDD78 pKa = 3.67IYY80 pKa = 11.5KK81 pKa = 10.48KK82 pKa = 10.6GSKK85 pKa = 9.63SDD87 pKa = 3.72YY88 pKa = 10.55SHH90 pKa = 7.7PCQAFQFCVSGNFSGKK106 pKa = 10.17RR107 pKa = 11.84FDD109 pKa = 3.75HH110 pKa = 6.1YY111 pKa = 11.25LYY113 pKa = 10.52GYY115 pKa = 7.99TITGFIDD122 pKa = 3.19IASSYY127 pKa = 9.19YY128 pKa = 10.61SGMSISTITLMPLQEE143 pKa = 4.48GSLNHH148 pKa = 7.2DD149 pKa = 3.9SEE151 pKa = 5.0EE152 pKa = 4.57DD153 pKa = 3.65CTTPPISTITQYY165 pKa = 11.05QRR167 pKa = 11.84IPEE170 pKa = 4.48PIIKK174 pKa = 9.55EE175 pKa = 3.91GCKK178 pKa = 9.75PVILQRR184 pKa = 11.84YY185 pKa = 8.53GEE187 pKa = 4.48SDD189 pKa = 4.02DD190 pKa = 4.39PTCIMYY196 pKa = 9.51WDD198 pKa = 4.04NTWDD202 pKa = 3.61NYY204 pKa = 10.87CDD206 pKa = 3.22VGFFNSQQRR215 pKa = 11.84DD216 pKa = 3.51HH217 pKa = 7.59DD218 pKa = 4.5PLVLPLNNYY227 pKa = 10.06LGISDD232 pKa = 4.97AFQDD236 pKa = 3.93FQSYY240 pKa = 8.5YY241 pKa = 10.68CKK243 pKa = 10.68SLDD246 pKa = 3.71LNQSYY251 pKa = 9.43SVCISIGEE259 pKa = 4.37TPTTVTYY266 pKa = 10.51HH267 pKa = 6.15SYY269 pKa = 11.69EE270 pKa = 4.32NITVNEE276 pKa = 4.14LLTRR280 pKa = 11.84IMILYY285 pKa = 10.37GEE287 pKa = 4.56EE288 pKa = 4.29KK289 pKa = 9.97VHH291 pKa = 6.77KK292 pKa = 10.53LPFRR296 pKa = 11.84NITIMAHH303 pKa = 5.56AQIQSLPLINSTCDD317 pKa = 3.31PNKK320 pKa = 10.27FDD322 pKa = 6.77DD323 pKa = 6.23DD324 pKa = 6.01DD325 pKa = 7.53DD326 pKa = 7.6DD327 pKa = 7.65DD328 pKa = 7.59DD329 pKa = 7.65DD330 pKa = 7.65DD331 pKa = 7.65DD332 pKa = 7.65DD333 pKa = 7.65DD334 pKa = 7.65DD335 pKa = 7.65DD336 pKa = 7.65DD337 pKa = 7.62DD338 pKa = 7.59DD339 pKa = 7.71DD340 pKa = 7.58DD341 pKa = 6.68DD342 pKa = 6.84DD343 pKa = 4.87EE344 pKa = 5.58YY345 pKa = 12.15NLYY348 pKa = 9.63TEE350 pKa = 4.42STPSKK355 pKa = 10.01VTTKK359 pKa = 10.03PKK361 pKa = 9.53KK362 pKa = 9.14TITDD366 pKa = 3.6EE367 pKa = 3.89YY368 pKa = 11.46DD369 pKa = 3.4SIFNSFDD376 pKa = 3.27NFDD379 pKa = 3.66LEE381 pKa = 4.61KK382 pKa = 10.78RR383 pKa = 3.75
MM1 pKa = 7.76IIYY4 pKa = 10.24IITLSLLFIKK14 pKa = 10.68VITSSKK20 pKa = 10.48EE21 pKa = 3.87PVKK24 pKa = 10.83NPQDD28 pKa = 3.13ILHH31 pKa = 6.73IIEE34 pKa = 4.42HH35 pKa = 6.23NKK37 pKa = 9.44TGVTACSLYY46 pKa = 11.02CFDD49 pKa = 4.71SSKK52 pKa = 11.34GLDD55 pKa = 3.42QPKK58 pKa = 8.87TFILPGKK65 pKa = 9.66YY66 pKa = 9.99SNNSIKK72 pKa = 10.87LEE74 pKa = 4.03VAIDD78 pKa = 3.67IYY80 pKa = 11.5KK81 pKa = 10.48KK82 pKa = 10.6GSKK85 pKa = 9.63SDD87 pKa = 3.72YY88 pKa = 10.55SHH90 pKa = 7.7PCQAFQFCVSGNFSGKK106 pKa = 10.17RR107 pKa = 11.84FDD109 pKa = 3.75HH110 pKa = 6.1YY111 pKa = 11.25LYY113 pKa = 10.52GYY115 pKa = 7.99TITGFIDD122 pKa = 3.19IASSYY127 pKa = 9.19YY128 pKa = 10.61SGMSISTITLMPLQEE143 pKa = 4.48GSLNHH148 pKa = 7.2DD149 pKa = 3.9SEE151 pKa = 5.0EE152 pKa = 4.57DD153 pKa = 3.65CTTPPISTITQYY165 pKa = 11.05QRR167 pKa = 11.84IPEE170 pKa = 4.48PIIKK174 pKa = 9.55EE175 pKa = 3.91GCKK178 pKa = 9.75PVILQRR184 pKa = 11.84YY185 pKa = 8.53GEE187 pKa = 4.48SDD189 pKa = 4.02DD190 pKa = 4.39PTCIMYY196 pKa = 9.51WDD198 pKa = 4.04NTWDD202 pKa = 3.61NYY204 pKa = 10.87CDD206 pKa = 3.22VGFFNSQQRR215 pKa = 11.84DD216 pKa = 3.51HH217 pKa = 7.59DD218 pKa = 4.5PLVLPLNNYY227 pKa = 10.06LGISDD232 pKa = 4.97AFQDD236 pKa = 3.93FQSYY240 pKa = 8.5YY241 pKa = 10.68CKK243 pKa = 10.68SLDD246 pKa = 3.71LNQSYY251 pKa = 9.43SVCISIGEE259 pKa = 4.37TPTTVTYY266 pKa = 10.51HH267 pKa = 6.15SYY269 pKa = 11.69EE270 pKa = 4.32NITVNEE276 pKa = 4.14LLTRR280 pKa = 11.84IMILYY285 pKa = 10.37GEE287 pKa = 4.56EE288 pKa = 4.29KK289 pKa = 9.97VHH291 pKa = 6.77KK292 pKa = 10.53LPFRR296 pKa = 11.84NITIMAHH303 pKa = 5.56AQIQSLPLINSTCDD317 pKa = 3.31PNKK320 pKa = 10.27FDD322 pKa = 6.77DD323 pKa = 6.23DD324 pKa = 6.01DD325 pKa = 7.53DD326 pKa = 7.6DD327 pKa = 7.65DD328 pKa = 7.59DD329 pKa = 7.65DD330 pKa = 7.65DD331 pKa = 7.65DD332 pKa = 7.65DD333 pKa = 7.65DD334 pKa = 7.65DD335 pKa = 7.65DD336 pKa = 7.65DD337 pKa = 7.62DD338 pKa = 7.59DD339 pKa = 7.71DD340 pKa = 7.58DD341 pKa = 6.68DD342 pKa = 6.84DD343 pKa = 4.87EE344 pKa = 5.58YY345 pKa = 12.15NLYY348 pKa = 9.63TEE350 pKa = 4.42STPSKK355 pKa = 10.01VTTKK359 pKa = 10.03PKK361 pKa = 9.53KK362 pKa = 9.14TITDD366 pKa = 3.6EE367 pKa = 3.89YY368 pKa = 11.46DD369 pKa = 3.4SIFNSFDD376 pKa = 3.27NFDD379 pKa = 3.66LEE381 pKa = 4.61KK382 pKa = 10.78RR383 pKa = 3.75
Molecular weight: 43.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A068EE70|A0A068EE70_9POXV DNA polymerase OS=Pigeonpox virus OX=10264 GN=fep_096 PE=3 SV=1
MM1 pKa = 7.54EE2 pKa = 5.37IARR5 pKa = 11.84EE6 pKa = 3.92TLITIGLTILVVVLVITGFSLVLRR30 pKa = 11.84LIPGVYY36 pKa = 9.71SAASRR41 pKa = 11.84SSFTAGKK48 pKa = 9.17VLRR51 pKa = 11.84FMEE54 pKa = 4.3IFSTVMFIPGIIILYY69 pKa = 9.17AAYY72 pKa = 9.46IRR74 pKa = 11.84KK75 pKa = 7.84TKK77 pKa = 9.57MKK79 pKa = 10.73NNN81 pKa = 3.69
MM1 pKa = 7.54EE2 pKa = 5.37IARR5 pKa = 11.84EE6 pKa = 3.92TLITIGLTILVVVLVITGFSLVLRR30 pKa = 11.84LIPGVYY36 pKa = 9.71SAASRR41 pKa = 11.84SSFTAGKK48 pKa = 9.17VLRR51 pKa = 11.84FMEE54 pKa = 4.3IFSTVMFIPGIIILYY69 pKa = 9.17AAYY72 pKa = 9.46IRR74 pKa = 11.84KK75 pKa = 7.84TKK77 pKa = 9.57MKK79 pKa = 10.73NNN81 pKa = 3.69
Molecular weight: 8.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
73067 |
34 |
1937 |
329.1 |
38.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.32 ± 0.105 | 2.203 ± 0.08 |
6.438 ± 0.119 | 5.674 ± 0.099 |
4.293 ± 0.123 | 3.861 ± 0.116 |
2.005 ± 0.058 | 10.274 ± 0.199 |
8.38 ± 0.14 | 9.193 ± 0.166 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.505 ± 0.057 | 7.344 ± 0.149 |
3.099 ± 0.106 | 1.99 ± 0.062 |
4.01 ± 0.083 | 7.768 ± 0.179 |
5.677 ± 0.118 | 5.637 ± 0.106 |
0.63 ± 0.039 | 5.698 ± 0.097 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
