
Plasmodium gonderi
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Plasmodium)
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
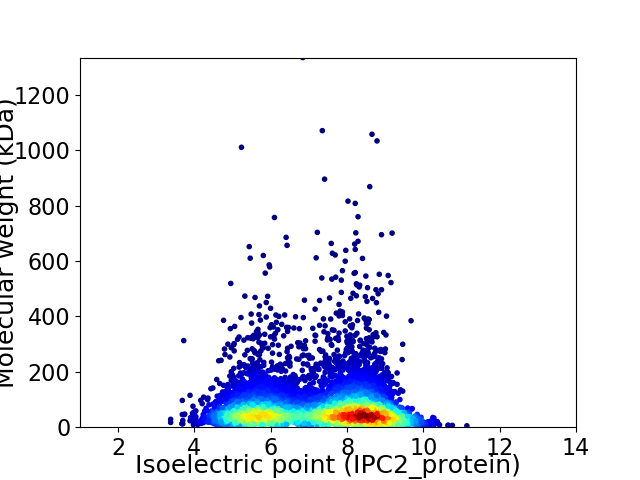
Virtual 2D-PAGE plot for 5915 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y1JIA4|A0A1Y1JIA4_PLAGO Non-specific serine/threonine protein kinase OS=Plasmodium gonderi OX=77519 GN=PGO_091160 PE=3 SV=1
MM1 pKa = 7.57EE2 pKa = 5.66EE3 pKa = 4.12GMSNNEE9 pKa = 3.94SSNGVLDD16 pKa = 4.23EE17 pKa = 4.59EE18 pKa = 4.54SQEE21 pKa = 4.16DD22 pKa = 4.2EE23 pKa = 4.68RR24 pKa = 11.84DD25 pKa = 3.28LSYY28 pKa = 10.65STFCNQEE35 pKa = 3.57EE36 pKa = 4.5GRR38 pKa = 11.84DD39 pKa = 3.58LSYY42 pKa = 11.62DD43 pKa = 3.48SFCNQEE49 pKa = 3.58EE50 pKa = 4.23RR51 pKa = 11.84GNISDD56 pKa = 3.46NSSYY60 pKa = 10.88NQEE63 pKa = 4.27EE64 pKa = 4.36EE65 pKa = 4.16IKK67 pKa = 10.06CQKK70 pKa = 10.8NEE72 pKa = 4.08DD73 pKa = 3.69HH74 pKa = 7.46DD75 pKa = 4.26GCANAGNYY83 pKa = 10.06LDD85 pKa = 5.46DD86 pKa = 4.67CNEE89 pKa = 3.9PKK91 pKa = 10.86GDD93 pKa = 4.03VRR95 pKa = 11.84DD96 pKa = 4.27KK97 pKa = 11.62EE98 pKa = 3.96NDD100 pKa = 3.14ADD102 pKa = 3.91GGQNEE107 pKa = 4.31EE108 pKa = 4.27EE109 pKa = 4.23EE110 pKa = 4.39EE111 pKa = 4.19EE112 pKa = 4.24EE113 pKa = 4.16EE114 pKa = 4.21EE115 pKa = 4.21EE116 pKa = 4.21EE117 pKa = 4.21EE118 pKa = 4.21EE119 pKa = 4.21EE120 pKa = 4.21EE121 pKa = 4.21EE122 pKa = 4.21EE123 pKa = 4.21EE124 pKa = 4.21EE125 pKa = 4.21EE126 pKa = 4.21EE127 pKa = 4.32EE128 pKa = 4.49EE129 pKa = 5.03EE130 pKa = 5.02EE131 pKa = 5.58EE132 pKa = 5.35EE133 pKa = 6.18DD134 pKa = 6.18DD135 pKa = 6.61DD136 pKa = 7.5DD137 pKa = 7.66DD138 pKa = 7.7DD139 pKa = 7.67DD140 pKa = 7.69DD141 pKa = 7.37DD142 pKa = 7.09DD143 pKa = 7.33DD144 pKa = 6.58YY145 pKa = 12.12DD146 pKa = 5.19DD147 pKa = 6.26DD148 pKa = 6.19EE149 pKa = 6.21YY150 pKa = 11.78DD151 pKa = 3.47EE152 pKa = 6.54DD153 pKa = 4.75DD154 pKa = 4.12FNEE157 pKa = 4.25ATVSWIEE164 pKa = 3.85WFCQLKK170 pKa = 10.52QNVFLVEE177 pKa = 3.73VDD179 pKa = 3.01EE180 pKa = 5.6DD181 pKa = 4.57FIRR184 pKa = 11.84DD185 pKa = 3.7EE186 pKa = 4.51FNLIGLQTKK195 pKa = 9.4VPHH198 pKa = 6.15FKK200 pKa = 10.65KK201 pKa = 10.34LLKK204 pKa = 10.33IILDD208 pKa = 4.07EE209 pKa = 6.84DD210 pKa = 5.47DD211 pKa = 6.35DD212 pKa = 7.41DD213 pKa = 7.56DD214 pKa = 7.67DD215 pKa = 7.63DD216 pKa = 7.14DD217 pKa = 6.94DD218 pKa = 6.04EE219 pKa = 6.22EE220 pKa = 6.6DD221 pKa = 6.07DD222 pKa = 4.29YY223 pKa = 12.15DD224 pKa = 5.36EE225 pKa = 6.96DD226 pKa = 5.22DD227 pKa = 5.26DD228 pKa = 4.99EE229 pKa = 5.65INRR232 pKa = 11.84GSDD235 pKa = 3.19EE236 pKa = 4.29LYY238 pKa = 10.5KK239 pKa = 11.25NKK241 pKa = 10.36DD242 pKa = 2.95IYY244 pKa = 9.74EE245 pKa = 4.04QNAACLYY252 pKa = 10.34GLIHH256 pKa = 6.74SRR258 pKa = 11.84FILTSKK264 pKa = 10.6GLALMRR270 pKa = 11.84EE271 pKa = 4.53KK272 pKa = 10.68YY273 pKa = 10.08KK274 pKa = 11.0SSIYY278 pKa = 8.53GTCPSIYY285 pKa = 9.91CDD287 pKa = 3.46NAKK290 pKa = 10.36LLPTAISEE298 pKa = 4.45VPKK301 pKa = 10.54FLSPLLYY308 pKa = 10.36CPRR311 pKa = 11.84CCEE314 pKa = 3.42TYY316 pKa = 10.3YY317 pKa = 10.09PHH319 pKa = 7.58KK320 pKa = 10.84NSLLNEE326 pKa = 4.14LDD328 pKa = 2.99GSYY331 pKa = 10.55FGTSFASFFALSFNIQSDD349 pKa = 3.52KK350 pKa = 11.0KK351 pKa = 10.21RR352 pKa = 11.84IYY354 pKa = 8.27YY355 pKa = 8.27TPQIFGFTINRR366 pKa = 11.84DD367 pKa = 2.7IRR369 pKa = 11.84EE370 pKa = 4.0NLFLDD375 pKa = 3.58MNKK378 pKa = 10.24EE379 pKa = 3.98NTEE382 pKa = 4.01SLEE385 pKa = 4.25EE386 pKa = 4.12YY387 pKa = 10.73SS388 pKa = 4.14
MM1 pKa = 7.57EE2 pKa = 5.66EE3 pKa = 4.12GMSNNEE9 pKa = 3.94SSNGVLDD16 pKa = 4.23EE17 pKa = 4.59EE18 pKa = 4.54SQEE21 pKa = 4.16DD22 pKa = 4.2EE23 pKa = 4.68RR24 pKa = 11.84DD25 pKa = 3.28LSYY28 pKa = 10.65STFCNQEE35 pKa = 3.57EE36 pKa = 4.5GRR38 pKa = 11.84DD39 pKa = 3.58LSYY42 pKa = 11.62DD43 pKa = 3.48SFCNQEE49 pKa = 3.58EE50 pKa = 4.23RR51 pKa = 11.84GNISDD56 pKa = 3.46NSSYY60 pKa = 10.88NQEE63 pKa = 4.27EE64 pKa = 4.36EE65 pKa = 4.16IKK67 pKa = 10.06CQKK70 pKa = 10.8NEE72 pKa = 4.08DD73 pKa = 3.69HH74 pKa = 7.46DD75 pKa = 4.26GCANAGNYY83 pKa = 10.06LDD85 pKa = 5.46DD86 pKa = 4.67CNEE89 pKa = 3.9PKK91 pKa = 10.86GDD93 pKa = 4.03VRR95 pKa = 11.84DD96 pKa = 4.27KK97 pKa = 11.62EE98 pKa = 3.96NDD100 pKa = 3.14ADD102 pKa = 3.91GGQNEE107 pKa = 4.31EE108 pKa = 4.27EE109 pKa = 4.23EE110 pKa = 4.39EE111 pKa = 4.19EE112 pKa = 4.24EE113 pKa = 4.16EE114 pKa = 4.21EE115 pKa = 4.21EE116 pKa = 4.21EE117 pKa = 4.21EE118 pKa = 4.21EE119 pKa = 4.21EE120 pKa = 4.21EE121 pKa = 4.21EE122 pKa = 4.21EE123 pKa = 4.21EE124 pKa = 4.21EE125 pKa = 4.21EE126 pKa = 4.21EE127 pKa = 4.32EE128 pKa = 4.49EE129 pKa = 5.03EE130 pKa = 5.02EE131 pKa = 5.58EE132 pKa = 5.35EE133 pKa = 6.18DD134 pKa = 6.18DD135 pKa = 6.61DD136 pKa = 7.5DD137 pKa = 7.66DD138 pKa = 7.7DD139 pKa = 7.67DD140 pKa = 7.69DD141 pKa = 7.37DD142 pKa = 7.09DD143 pKa = 7.33DD144 pKa = 6.58YY145 pKa = 12.12DD146 pKa = 5.19DD147 pKa = 6.26DD148 pKa = 6.19EE149 pKa = 6.21YY150 pKa = 11.78DD151 pKa = 3.47EE152 pKa = 6.54DD153 pKa = 4.75DD154 pKa = 4.12FNEE157 pKa = 4.25ATVSWIEE164 pKa = 3.85WFCQLKK170 pKa = 10.52QNVFLVEE177 pKa = 3.73VDD179 pKa = 3.01EE180 pKa = 5.6DD181 pKa = 4.57FIRR184 pKa = 11.84DD185 pKa = 3.7EE186 pKa = 4.51FNLIGLQTKK195 pKa = 9.4VPHH198 pKa = 6.15FKK200 pKa = 10.65KK201 pKa = 10.34LLKK204 pKa = 10.33IILDD208 pKa = 4.07EE209 pKa = 6.84DD210 pKa = 5.47DD211 pKa = 6.35DD212 pKa = 7.41DD213 pKa = 7.56DD214 pKa = 7.67DD215 pKa = 7.63DD216 pKa = 7.14DD217 pKa = 6.94DD218 pKa = 6.04EE219 pKa = 6.22EE220 pKa = 6.6DD221 pKa = 6.07DD222 pKa = 4.29YY223 pKa = 12.15DD224 pKa = 5.36EE225 pKa = 6.96DD226 pKa = 5.22DD227 pKa = 5.26DD228 pKa = 4.99EE229 pKa = 5.65INRR232 pKa = 11.84GSDD235 pKa = 3.19EE236 pKa = 4.29LYY238 pKa = 10.5KK239 pKa = 11.25NKK241 pKa = 10.36DD242 pKa = 2.95IYY244 pKa = 9.74EE245 pKa = 4.04QNAACLYY252 pKa = 10.34GLIHH256 pKa = 6.74SRR258 pKa = 11.84FILTSKK264 pKa = 10.6GLALMRR270 pKa = 11.84EE271 pKa = 4.53KK272 pKa = 10.68YY273 pKa = 10.08KK274 pKa = 11.0SSIYY278 pKa = 8.53GTCPSIYY285 pKa = 9.91CDD287 pKa = 3.46NAKK290 pKa = 10.36LLPTAISEE298 pKa = 4.45VPKK301 pKa = 10.54FLSPLLYY308 pKa = 10.36CPRR311 pKa = 11.84CCEE314 pKa = 3.42TYY316 pKa = 10.3YY317 pKa = 10.09PHH319 pKa = 7.58KK320 pKa = 10.84NSLLNEE326 pKa = 4.14LDD328 pKa = 2.99GSYY331 pKa = 10.55FGTSFASFFALSFNIQSDD349 pKa = 3.52KK350 pKa = 11.0KK351 pKa = 10.21RR352 pKa = 11.84IYY354 pKa = 8.27YY355 pKa = 8.27TPQIFGFTINRR366 pKa = 11.84DD367 pKa = 2.7IRR369 pKa = 11.84EE370 pKa = 4.0NLFLDD375 pKa = 3.58MNKK378 pKa = 10.24EE379 pKa = 3.98NTEE382 pKa = 4.01SLEE385 pKa = 4.25EE386 pKa = 4.12YY387 pKa = 10.73SS388 pKa = 4.14
Molecular weight: 45.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y1JIP1|A0A1Y1JIP1_PLAGO TBC domain protein OS=Plasmodium gonderi OX=77519 GN=PGO_120820 PE=4 SV=1
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4048994 |
36 |
11537 |
684.5 |
79.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.865 ± 0.02 | 2.006 ± 0.015 |
5.666 ± 0.022 | 7.838 ± 0.045 |
4.634 ± 0.028 | 4.007 ± 0.029 |
2.499 ± 0.012 | 7.934 ± 0.036 |
10.839 ± 0.037 | 7.895 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.34 ± 0.014 | 10.377 ± 0.054 |
2.308 ± 0.017 | 2.904 ± 0.014 |
3.761 ± 0.023 | 7.703 ± 0.027 |
4.523 ± 0.016 | 4.469 ± 0.016 |
0.579 ± 0.007 | 4.852 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
