
Aquimarina amphilecti
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aquimarina
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
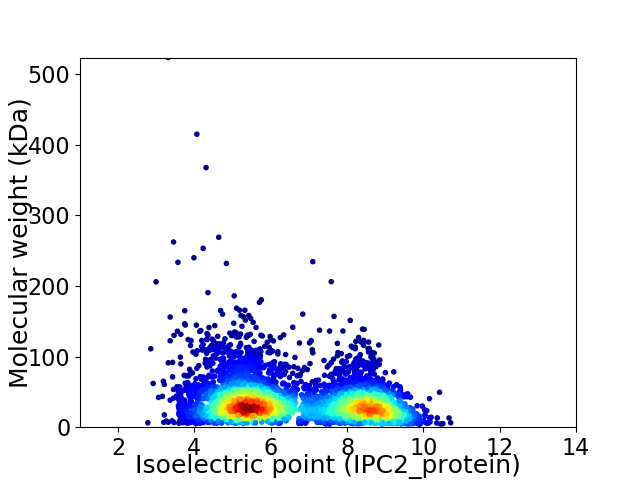
Virtual 2D-PAGE plot for 4646 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
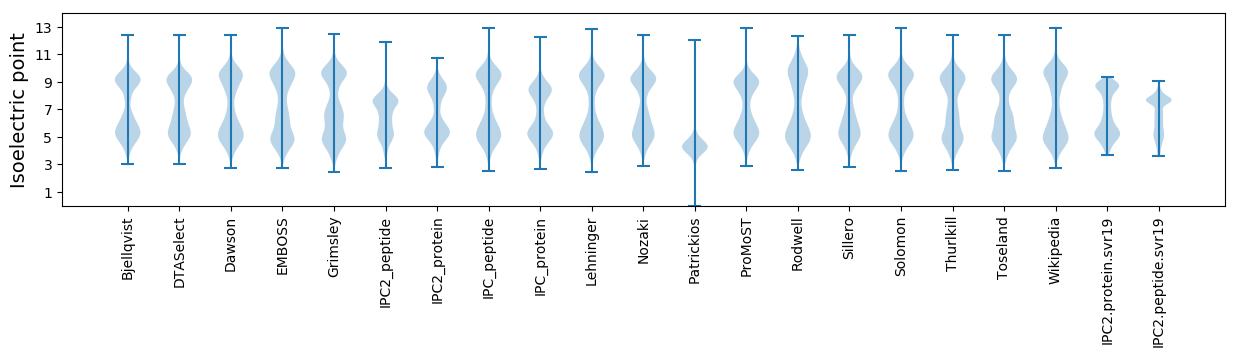
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H7Q8U0|A0A1H7Q8U0_9FLAO CubicO group peptidase beta-lactamase class C family OS=Aquimarina amphilecti OX=1038014 GN=SAMN04487910_2452 PE=4 SV=1
MM1 pKa = 7.26NKK3 pKa = 9.86IITFLIFVCTISITAQNSYY22 pKa = 10.21RR23 pKa = 11.84INYY26 pKa = 7.53QAVVRR31 pKa = 11.84DD32 pKa = 3.57AGNIVVGDD40 pKa = 3.99TNVGVQINILQTSASGTSVYY60 pKa = 10.82SEE62 pKa = 4.12NQSVTTNINGLASLVIGNAAGFDD85 pKa = 4.09SIDD88 pKa = 3.5WSAGPYY94 pKa = 9.83FIRR97 pKa = 11.84IEE99 pKa = 3.97IDD101 pKa = 3.15PAGGTSYY108 pKa = 10.23TLSTVNQLLSVPYY121 pKa = 9.55ALHH124 pKa = 6.69SATADD129 pKa = 3.3VAATANYY136 pKa = 10.41NSLTNLPTIITPAQAAKK153 pKa = 10.04IDD155 pKa = 4.62FITVTSAVNIDD166 pKa = 3.56QLQTDD171 pKa = 3.95VTANNAKK178 pKa = 8.65VTFPGFGTVPGTALEE193 pKa = 4.29GDD195 pKa = 3.57NVIWTKK201 pKa = 11.56SMTDD205 pKa = 3.5DD206 pKa = 3.11IFYY209 pKa = 10.65NAGNVGIGVDD219 pKa = 3.9EE220 pKa = 4.54SSSFSGSKK228 pKa = 9.99LHH230 pKa = 6.55VGGPILFDD238 pKa = 3.29GTPAATVPGSLYY250 pKa = 10.69YY251 pKa = 10.89DD252 pKa = 3.47NTGSGSFHH260 pKa = 7.03YY261 pKa = 10.17IDD263 pKa = 3.77NTNTDD268 pKa = 3.13VTINTGAVTYY278 pKa = 10.7AGGLWSIEE286 pKa = 3.98NGDD289 pKa = 3.78AVISNDD295 pKa = 3.35VFIQNSLGVGQDD307 pKa = 2.43ISTGVDD313 pKa = 3.8FGFNTLLLAEE323 pKa = 4.41NNLRR327 pKa = 11.84ILFDD331 pKa = 4.83DD332 pKa = 4.26SDD334 pKa = 5.25DD335 pKa = 4.05ISGTLPANDD344 pKa = 3.39WQIEE348 pKa = 4.19INEE351 pKa = 4.33TANGGTSHH359 pKa = 7.2FAIVDD364 pKa = 3.59VTGATTPFKK373 pKa = 10.43ILAGAAEE380 pKa = 4.36HH381 pKa = 6.43SLFVATNGNVGLGTNNPTNKK401 pKa = 10.14LEE403 pKa = 4.09VVGAIKK409 pKa = 10.59ADD411 pKa = 3.68SFIGDD416 pKa = 3.53GSGITGIATGTGGVTNAGDD435 pKa = 3.47TTIAADD441 pKa = 3.72NDD443 pKa = 3.68ADD445 pKa = 4.31TVGEE449 pKa = 4.52IAFQTQNITRR459 pKa = 11.84MTVSNTGFIGIGTTTPSVALDD480 pKa = 3.66VIGDD484 pKa = 3.82AKK486 pKa = 10.29VDD488 pKa = 3.5NMTLNGNASVQSIVITPNVEE508 pKa = 3.99TSATTFPTTYY518 pKa = 10.95DD519 pKa = 3.39VTDD522 pKa = 3.53KK523 pKa = 11.6SFVTLDD529 pKa = 3.36GQAASSIQAFSGGITGQSIIVTALTSTITLEE560 pKa = 4.38HH561 pKa = 6.31NAGTAAQNFQLAGNANIILTANSSATFVYY590 pKa = 10.55DD591 pKa = 3.18GTNWYY596 pKa = 10.15CVGLNNN602 pKa = 5.33
MM1 pKa = 7.26NKK3 pKa = 9.86IITFLIFVCTISITAQNSYY22 pKa = 10.21RR23 pKa = 11.84INYY26 pKa = 7.53QAVVRR31 pKa = 11.84DD32 pKa = 3.57AGNIVVGDD40 pKa = 3.99TNVGVQINILQTSASGTSVYY60 pKa = 10.82SEE62 pKa = 4.12NQSVTTNINGLASLVIGNAAGFDD85 pKa = 4.09SIDD88 pKa = 3.5WSAGPYY94 pKa = 9.83FIRR97 pKa = 11.84IEE99 pKa = 3.97IDD101 pKa = 3.15PAGGTSYY108 pKa = 10.23TLSTVNQLLSVPYY121 pKa = 9.55ALHH124 pKa = 6.69SATADD129 pKa = 3.3VAATANYY136 pKa = 10.41NSLTNLPTIITPAQAAKK153 pKa = 10.04IDD155 pKa = 4.62FITVTSAVNIDD166 pKa = 3.56QLQTDD171 pKa = 3.95VTANNAKK178 pKa = 8.65VTFPGFGTVPGTALEE193 pKa = 4.29GDD195 pKa = 3.57NVIWTKK201 pKa = 11.56SMTDD205 pKa = 3.5DD206 pKa = 3.11IFYY209 pKa = 10.65NAGNVGIGVDD219 pKa = 3.9EE220 pKa = 4.54SSSFSGSKK228 pKa = 9.99LHH230 pKa = 6.55VGGPILFDD238 pKa = 3.29GTPAATVPGSLYY250 pKa = 10.69YY251 pKa = 10.89DD252 pKa = 3.47NTGSGSFHH260 pKa = 7.03YY261 pKa = 10.17IDD263 pKa = 3.77NTNTDD268 pKa = 3.13VTINTGAVTYY278 pKa = 10.7AGGLWSIEE286 pKa = 3.98NGDD289 pKa = 3.78AVISNDD295 pKa = 3.35VFIQNSLGVGQDD307 pKa = 2.43ISTGVDD313 pKa = 3.8FGFNTLLLAEE323 pKa = 4.41NNLRR327 pKa = 11.84ILFDD331 pKa = 4.83DD332 pKa = 4.26SDD334 pKa = 5.25DD335 pKa = 4.05ISGTLPANDD344 pKa = 3.39WQIEE348 pKa = 4.19INEE351 pKa = 4.33TANGGTSHH359 pKa = 7.2FAIVDD364 pKa = 3.59VTGATTPFKK373 pKa = 10.43ILAGAAEE380 pKa = 4.36HH381 pKa = 6.43SLFVATNGNVGLGTNNPTNKK401 pKa = 10.14LEE403 pKa = 4.09VVGAIKK409 pKa = 10.59ADD411 pKa = 3.68SFIGDD416 pKa = 3.53GSGITGIATGTGGVTNAGDD435 pKa = 3.47TTIAADD441 pKa = 3.72NDD443 pKa = 3.68ADD445 pKa = 4.31TVGEE449 pKa = 4.52IAFQTQNITRR459 pKa = 11.84MTVSNTGFIGIGTTTPSVALDD480 pKa = 3.66VIGDD484 pKa = 3.82AKK486 pKa = 10.29VDD488 pKa = 3.5NMTLNGNASVQSIVITPNVEE508 pKa = 3.99TSATTFPTTYY518 pKa = 10.95DD519 pKa = 3.39VTDD522 pKa = 3.53KK523 pKa = 11.6SFVTLDD529 pKa = 3.36GQAASSIQAFSGGITGQSIIVTALTSTITLEE560 pKa = 4.38HH561 pKa = 6.31NAGTAAQNFQLAGNANIILTANSSATFVYY590 pKa = 10.55DD591 pKa = 3.18GTNWYY596 pKa = 10.15CVGLNNN602 pKa = 5.33
Molecular weight: 62.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H7QFH8|A0A1H7QFH8_9FLAO Glutamate 5-kinase OS=Aquimarina amphilecti OX=1038014 GN=proB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 5.43KK51 pKa = 10.29HH52 pKa = 5.04
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 5.43KK51 pKa = 10.29HH52 pKa = 5.04
Molecular weight: 6.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1587142 |
36 |
4991 |
341.6 |
38.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.603 ± 0.037 | 0.751 ± 0.011 |
5.799 ± 0.033 | 6.552 ± 0.032 |
5.238 ± 0.032 | 6.407 ± 0.039 |
1.652 ± 0.015 | 8.522 ± 0.041 |
7.825 ± 0.057 | 9.075 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.012 ± 0.015 | 6.485 ± 0.033 |
3.26 ± 0.023 | 3.297 ± 0.019 |
3.381 ± 0.022 | 6.905 ± 0.029 |
5.971 ± 0.041 | 5.994 ± 0.032 |
1.085 ± 0.014 | 4.185 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
