
Bacillus sp. P16(2019)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; unclassified Bacillus (in: Bacteria)
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
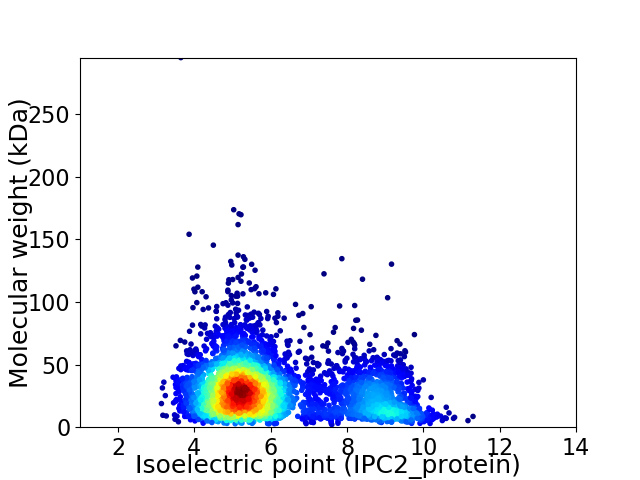
Virtual 2D-PAGE plot for 4393 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A554A316|A0A554A316_9BACI GAF domain-containing protein OS=Bacillus sp. P16(2019) OX=2597270 GN=FN960_00485 PE=4 SV=1
MM1 pKa = 7.29NLSNKK6 pKa = 8.5WFVGVSLSAIVLVTGCSNEE25 pKa = 3.7GSGDD29 pKa = 3.57GDD31 pKa = 3.65SDD33 pKa = 3.44QTTIRR38 pKa = 11.84VHH40 pKa = 4.99HH41 pKa = 7.26WYY43 pKa = 10.57NEE45 pKa = 4.06EE46 pKa = 3.54QDD48 pKa = 3.33NWDD51 pKa = 3.74EE52 pKa = 4.02VFAAYY57 pKa = 9.97EE58 pKa = 4.15EE59 pKa = 4.28EE60 pKa = 4.03TGIKK64 pKa = 9.46IEE66 pKa = 4.74SVTPEE71 pKa = 3.92NNDD74 pKa = 3.35ANEE77 pKa = 4.02TLQQIDD83 pKa = 3.62LAAASGDD90 pKa = 3.63QLDD93 pKa = 4.21VIMVNDD99 pKa = 3.93AANYY103 pKa = 7.2SQRR106 pKa = 11.84ISQGMFEE113 pKa = 4.8PLNDD117 pKa = 3.86YY118 pKa = 10.92LDD120 pKa = 3.97EE121 pKa = 6.59AGIQYY126 pKa = 10.85EE127 pKa = 4.88DD128 pKa = 5.2DD129 pKa = 3.98YY130 pKa = 12.1QNDD133 pKa = 3.77TSVDD137 pKa = 3.33NQYY140 pKa = 11.13YY141 pKa = 10.69ALPGKK146 pKa = 9.84YY147 pKa = 9.11NQSFVMLNEE156 pKa = 4.52DD157 pKa = 3.91ALDD160 pKa = 3.92EE161 pKa = 4.95AGLDD165 pKa = 4.45IPVDD169 pKa = 3.98WTWDD173 pKa = 3.36DD174 pKa = 3.65YY175 pKa = 11.5MDD177 pKa = 3.9YY178 pKa = 11.53AEE180 pKa = 5.62ALTEE184 pKa = 4.28GEE186 pKa = 4.57GSSKK190 pKa = 10.63RR191 pKa = 11.84YY192 pKa = 8.39GTFFHH197 pKa = 6.25TWVDD201 pKa = 3.99YY202 pKa = 11.5VKK204 pKa = 10.4LAAMNQSEE212 pKa = 4.05QSNLVQDD219 pKa = 4.94DD220 pKa = 4.46GVTSNINSDD229 pKa = 4.25IIRR232 pKa = 11.84QSLEE236 pKa = 3.14IRR238 pKa = 11.84QRR240 pKa = 11.84GIDD243 pKa = 3.63EE244 pKa = 4.85GSATPHH250 pKa = 7.16ADD252 pKa = 3.85TISLDD257 pKa = 3.62LNYY260 pKa = 10.26RR261 pKa = 11.84PQFFSEE267 pKa = 4.29SAAMIMTGSWMINEE281 pKa = 4.16AGGRR285 pKa = 11.84PEE287 pKa = 3.94EE288 pKa = 4.34PSTFRR293 pKa = 11.84TAFAPYY299 pKa = 9.78PKK301 pKa = 10.76ANDD304 pKa = 3.56NDD306 pKa = 5.54PITTPAGLDD315 pKa = 3.45FLTVYY320 pKa = 10.41SGSDD324 pKa = 3.15HH325 pKa = 6.19KK326 pKa = 11.36QEE328 pKa = 4.31AFDD331 pKa = 4.41FIRR334 pKa = 11.84WYY336 pKa = 6.68TTEE339 pKa = 5.24GILDD343 pKa = 3.39QGMYY347 pKa = 10.23YY348 pKa = 9.83PDD350 pKa = 4.17YY351 pKa = 10.95KK352 pKa = 11.05NADD355 pKa = 3.27IDD357 pKa = 4.23TVLDD361 pKa = 3.72NLLDD365 pKa = 3.68NTPNIEE371 pKa = 4.11MVNRR375 pKa = 11.84DD376 pKa = 3.48SLQATLEE383 pKa = 4.11ASEE386 pKa = 4.46PADD389 pKa = 3.37MNIPVPYY396 pKa = 9.37IGEE399 pKa = 4.19VVTAYY404 pKa = 10.26MNEE407 pKa = 3.9TDD409 pKa = 4.18RR410 pKa = 11.84FLLEE414 pKa = 4.2EE415 pKa = 4.18QDD417 pKa = 4.32LEE419 pKa = 4.58TTIEE423 pKa = 3.91RR424 pKa = 11.84AEE426 pKa = 4.17EE427 pKa = 4.12SVQQVIDD434 pKa = 3.59NNANN438 pKa = 3.09
MM1 pKa = 7.29NLSNKK6 pKa = 8.5WFVGVSLSAIVLVTGCSNEE25 pKa = 3.7GSGDD29 pKa = 3.57GDD31 pKa = 3.65SDD33 pKa = 3.44QTTIRR38 pKa = 11.84VHH40 pKa = 4.99HH41 pKa = 7.26WYY43 pKa = 10.57NEE45 pKa = 4.06EE46 pKa = 3.54QDD48 pKa = 3.33NWDD51 pKa = 3.74EE52 pKa = 4.02VFAAYY57 pKa = 9.97EE58 pKa = 4.15EE59 pKa = 4.28EE60 pKa = 4.03TGIKK64 pKa = 9.46IEE66 pKa = 4.74SVTPEE71 pKa = 3.92NNDD74 pKa = 3.35ANEE77 pKa = 4.02TLQQIDD83 pKa = 3.62LAAASGDD90 pKa = 3.63QLDD93 pKa = 4.21VIMVNDD99 pKa = 3.93AANYY103 pKa = 7.2SQRR106 pKa = 11.84ISQGMFEE113 pKa = 4.8PLNDD117 pKa = 3.86YY118 pKa = 10.92LDD120 pKa = 3.97EE121 pKa = 6.59AGIQYY126 pKa = 10.85EE127 pKa = 4.88DD128 pKa = 5.2DD129 pKa = 3.98YY130 pKa = 12.1QNDD133 pKa = 3.77TSVDD137 pKa = 3.33NQYY140 pKa = 11.13YY141 pKa = 10.69ALPGKK146 pKa = 9.84YY147 pKa = 9.11NQSFVMLNEE156 pKa = 4.52DD157 pKa = 3.91ALDD160 pKa = 3.92EE161 pKa = 4.95AGLDD165 pKa = 4.45IPVDD169 pKa = 3.98WTWDD173 pKa = 3.36DD174 pKa = 3.65YY175 pKa = 11.5MDD177 pKa = 3.9YY178 pKa = 11.53AEE180 pKa = 5.62ALTEE184 pKa = 4.28GEE186 pKa = 4.57GSSKK190 pKa = 10.63RR191 pKa = 11.84YY192 pKa = 8.39GTFFHH197 pKa = 6.25TWVDD201 pKa = 3.99YY202 pKa = 11.5VKK204 pKa = 10.4LAAMNQSEE212 pKa = 4.05QSNLVQDD219 pKa = 4.94DD220 pKa = 4.46GVTSNINSDD229 pKa = 4.25IIRR232 pKa = 11.84QSLEE236 pKa = 3.14IRR238 pKa = 11.84QRR240 pKa = 11.84GIDD243 pKa = 3.63EE244 pKa = 4.85GSATPHH250 pKa = 7.16ADD252 pKa = 3.85TISLDD257 pKa = 3.62LNYY260 pKa = 10.26RR261 pKa = 11.84PQFFSEE267 pKa = 4.29SAAMIMTGSWMINEE281 pKa = 4.16AGGRR285 pKa = 11.84PEE287 pKa = 3.94EE288 pKa = 4.34PSTFRR293 pKa = 11.84TAFAPYY299 pKa = 9.78PKK301 pKa = 10.76ANDD304 pKa = 3.56NDD306 pKa = 5.54PITTPAGLDD315 pKa = 3.45FLTVYY320 pKa = 10.41SGSDD324 pKa = 3.15HH325 pKa = 6.19KK326 pKa = 11.36QEE328 pKa = 4.31AFDD331 pKa = 4.41FIRR334 pKa = 11.84WYY336 pKa = 6.68TTEE339 pKa = 5.24GILDD343 pKa = 3.39QGMYY347 pKa = 10.23YY348 pKa = 9.83PDD350 pKa = 4.17YY351 pKa = 10.95KK352 pKa = 11.05NADD355 pKa = 3.27IDD357 pKa = 4.23TVLDD361 pKa = 3.72NLLDD365 pKa = 3.68NTPNIEE371 pKa = 4.11MVNRR375 pKa = 11.84DD376 pKa = 3.48SLQATLEE383 pKa = 4.11ASEE386 pKa = 4.46PADD389 pKa = 3.37MNIPVPYY396 pKa = 9.37IGEE399 pKa = 4.19VVTAYY404 pKa = 10.26MNEE407 pKa = 3.9TDD409 pKa = 4.18RR410 pKa = 11.84FLLEE414 pKa = 4.2EE415 pKa = 4.18QDD417 pKa = 4.32LEE419 pKa = 4.58TTIEE423 pKa = 3.91RR424 pKa = 11.84AEE426 pKa = 4.17EE427 pKa = 4.12SVQQVIDD434 pKa = 3.59NNANN438 pKa = 3.09
Molecular weight: 49.26 kDa
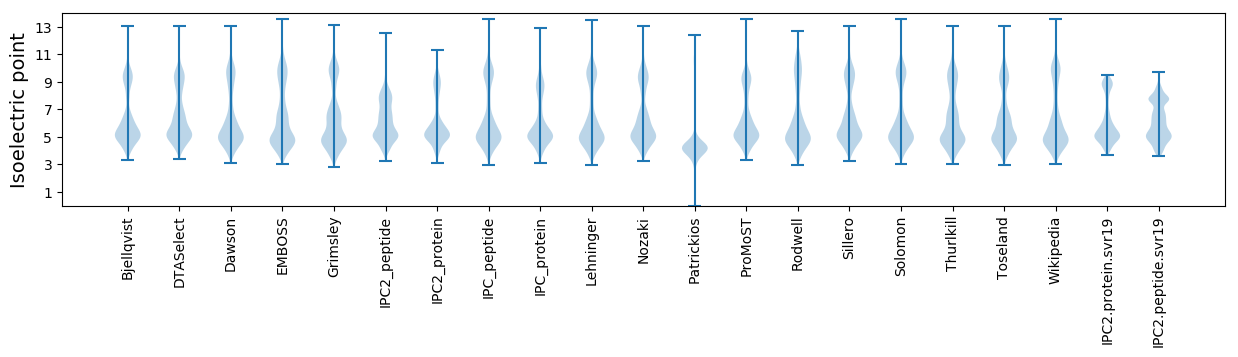
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A553ZSJ3|A0A553ZSJ3_9BACI EAL domain-containing protein (Fragment) OS=Bacillus sp. P16(2019) OX=2597270 GN=FN960_21955 PE=4 SV=1
RR1 pKa = 7.24ARR3 pKa = 11.84PVRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84SGSRR12 pKa = 11.84GARR15 pKa = 11.84RR16 pKa = 11.84PGRR19 pKa = 11.84AAGAPSSARR28 pKa = 11.84TRR30 pKa = 11.84RR31 pKa = 11.84PAPAAPPRR39 pKa = 11.84GAPGRR44 pKa = 11.84PAPRR48 pKa = 11.84WSAPAGSGRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84AGRR62 pKa = 11.84PSRR65 pKa = 11.84PGPGARR71 pKa = 11.84PRR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84GRR77 pKa = 11.84CGRR80 pKa = 11.84TGG82 pKa = 2.91
RR1 pKa = 7.24ARR3 pKa = 11.84PVRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84SGSRR12 pKa = 11.84GARR15 pKa = 11.84RR16 pKa = 11.84PGRR19 pKa = 11.84AAGAPSSARR28 pKa = 11.84TRR30 pKa = 11.84RR31 pKa = 11.84PAPAAPPRR39 pKa = 11.84GAPGRR44 pKa = 11.84PAPRR48 pKa = 11.84WSAPAGSGRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84AGRR62 pKa = 11.84PSRR65 pKa = 11.84PGPGARR71 pKa = 11.84PRR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84GRR77 pKa = 11.84CGRR80 pKa = 11.84TGG82 pKa = 2.91
Molecular weight: 8.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1227302 |
21 |
2698 |
279.4 |
31.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.289 ± 0.04 | 0.673 ± 0.011 |
5.201 ± 0.033 | 7.659 ± 0.045 |
4.409 ± 0.035 | 6.853 ± 0.039 |
2.284 ± 0.02 | 7.41 ± 0.034 |
5.678 ± 0.038 | 10.139 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.754 ± 0.019 | 3.984 ± 0.031 |
3.642 ± 0.022 | 4.219 ± 0.028 |
4.207 ± 0.032 | 6.373 ± 0.027 |
5.665 ± 0.024 | 7.01 ± 0.031 |
1.097 ± 0.014 | 3.453 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
