
Synechococcus phage S-SM1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Thetisvirus; Synechococcus virus SSM1
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
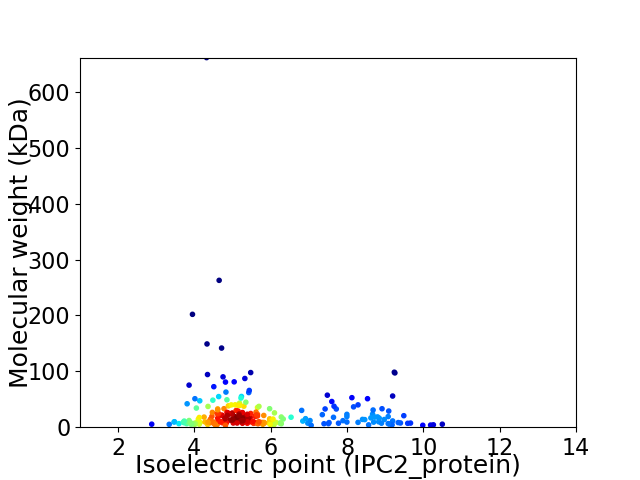
Virtual 2D-PAGE plot for 234 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
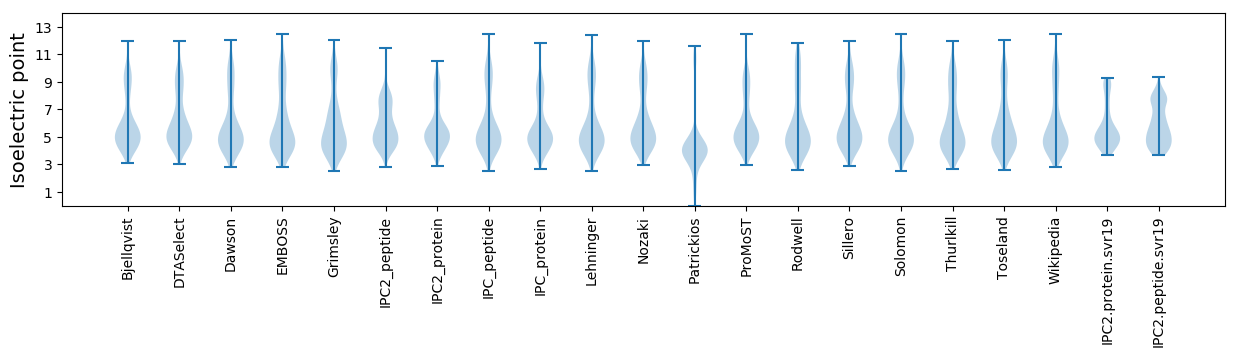
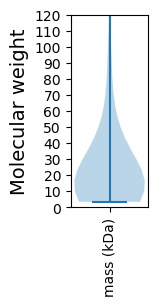
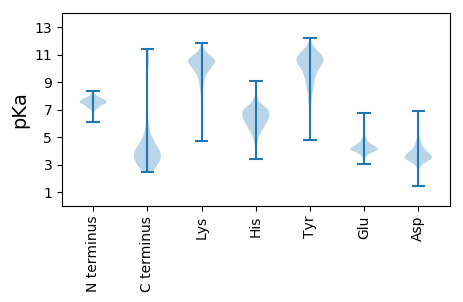
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E3SIN4|E3SIN4_9CAUD Uncharacterized protein OS=Synechococcus phage S-SM1 OX=444859 GN=SSM1_228 PE=4 SV=1
MM1 pKa = 7.74AYY3 pKa = 9.79YY4 pKa = 10.27YY5 pKa = 9.95PEE7 pKa = 4.45GAFGPICDD15 pKa = 5.26LPLTDD20 pKa = 4.82AQIIEE25 pKa = 4.3QGRR28 pKa = 11.84SAISIDD34 pKa = 3.29EE35 pKa = 4.14GLIEE39 pKa = 4.2QYY41 pKa = 11.21GVVPGIYY48 pKa = 10.17DD49 pKa = 3.46EE50 pKa = 6.08FLDD53 pKa = 4.42FWGEE57 pKa = 3.78FNFPVFLRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.33CKK69 pKa = 10.21EE70 pKa = 3.65RR71 pKa = 11.84VLDD74 pKa = 4.65DD75 pKa = 3.47GTIEE79 pKa = 5.33KK80 pKa = 9.97FDD82 pKa = 4.15CVDD85 pKa = 3.32EE86 pKa = 4.3YY87 pKa = 11.73SRR89 pKa = 11.84PLPTSQEE96 pKa = 4.33SFVSATDD103 pKa = 4.47DD104 pKa = 3.9IGLKK108 pKa = 10.7DD109 pKa = 4.57LFFEE113 pKa = 4.8PKK115 pKa = 8.91LTPEE119 pKa = 3.99TCSPYY124 pKa = 10.28QADD127 pKa = 3.42INIRR131 pKa = 11.84PLKK134 pKa = 10.33FFGPNGVLIKK144 pKa = 9.47RR145 pKa = 11.84TPVEE149 pKa = 3.83GSTPVTFPVTSEE161 pKa = 4.0NQSIPNNATITATFSEE177 pKa = 4.54DD178 pKa = 3.15RR179 pKa = 11.84QNLVIGGTGEE189 pKa = 4.76GIVQLRR195 pKa = 11.84FTWDD199 pKa = 3.58DD200 pKa = 3.53KK201 pKa = 11.52PSISGLAVGTLTVAGASFTQSGEE224 pKa = 4.01KK225 pKa = 10.81GSDD228 pKa = 3.28NDD230 pKa = 3.61STSIVTAGQSYY241 pKa = 9.44PITLSGNSGTSGSRR255 pKa = 11.84LQGNTQIEE263 pKa = 4.39YY264 pKa = 10.8DD265 pKa = 3.87DD266 pKa = 5.03DD267 pKa = 4.52VPGFDD272 pKa = 4.21VNATLSITEE281 pKa = 4.12VLPTTTSTNVAGYY294 pKa = 9.57WSDD297 pKa = 3.28EE298 pKa = 3.96GNKK301 pKa = 10.01YY302 pKa = 9.91GVWVNPAICTLPQQPQSVSYY322 pKa = 10.54KK323 pKa = 9.09ITIDD327 pKa = 3.42EE328 pKa = 4.48ADD330 pKa = 3.35TYY332 pKa = 11.46GFTFGCDD339 pKa = 3.2DD340 pKa = 4.35NATLTINDD348 pKa = 3.73EE349 pKa = 4.15TSAFLTAVGGIFEE362 pKa = 4.7GGSYY366 pKa = 7.55NTPYY370 pKa = 10.26TGTRR374 pKa = 11.84ALTAGTLILTVNCTNSDD391 pKa = 4.06AGFQDD396 pKa = 4.48ANGDD400 pKa = 4.03PTGLAYY406 pKa = 10.23DD407 pKa = 3.89WQRR410 pKa = 11.84NPGGWYY416 pKa = 9.74IKK418 pKa = 8.67MCKK421 pKa = 10.03GGLCASGTNINWVPVGPHH439 pKa = 6.26PAWSDD444 pKa = 3.58FMDD447 pKa = 4.81EE448 pKa = 4.32YY449 pKa = 11.67AVFPSNIDD457 pKa = 3.41PLLDD461 pKa = 3.32QTQQATWNINIPTTGNYY478 pKa = 8.38TFEE481 pKa = 4.57CSADD485 pKa = 3.48NNATFTLDD493 pKa = 3.08GTQIATSNSFTTTTSVSLTNISEE516 pKa = 4.57GPHH519 pKa = 5.19TLVVSVFNGTNSNGANEE536 pKa = 4.21WTTNPGGVAWRR547 pKa = 11.84IQYY550 pKa = 10.69GGGSIDD556 pKa = 5.95ANFQSNGRR564 pKa = 11.84LLVTGAGAARR574 pKa = 11.84LDD576 pKa = 5.42FDD578 pKa = 5.96FEE580 pKa = 4.43WDD582 pKa = 4.53DD583 pKa = 4.11NPSTYY588 pKa = 9.85DD589 pKa = 3.25TALGTVAYY597 pKa = 7.31PQLGVSFTQNTGSSSGSDD615 pKa = 2.92SDD617 pKa = 3.82NVNNVTAGDD626 pKa = 3.83YY627 pKa = 11.17GVTILNNSGGFTVEE641 pKa = 3.78NSGKK645 pKa = 9.95RR646 pKa = 11.84LCFKK650 pKa = 10.92DD651 pKa = 5.35LDD653 pKa = 4.46GNDD656 pKa = 4.12CNAQLDD662 pKa = 4.0LSIVQGDD669 pKa = 3.66ATIASSLDD677 pKa = 3.54LNTPGDD683 pKa = 4.26GNLFWHH689 pKa = 6.4TRR691 pKa = 11.84LATGYY696 pKa = 8.63TFLEE700 pKa = 4.92DD701 pKa = 3.27NN702 pKa = 4.34
MM1 pKa = 7.74AYY3 pKa = 9.79YY4 pKa = 10.27YY5 pKa = 9.95PEE7 pKa = 4.45GAFGPICDD15 pKa = 5.26LPLTDD20 pKa = 4.82AQIIEE25 pKa = 4.3QGRR28 pKa = 11.84SAISIDD34 pKa = 3.29EE35 pKa = 4.14GLIEE39 pKa = 4.2QYY41 pKa = 11.21GVVPGIYY48 pKa = 10.17DD49 pKa = 3.46EE50 pKa = 6.08FLDD53 pKa = 4.42FWGEE57 pKa = 3.78FNFPVFLRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.33CKK69 pKa = 10.21EE70 pKa = 3.65RR71 pKa = 11.84VLDD74 pKa = 4.65DD75 pKa = 3.47GTIEE79 pKa = 5.33KK80 pKa = 9.97FDD82 pKa = 4.15CVDD85 pKa = 3.32EE86 pKa = 4.3YY87 pKa = 11.73SRR89 pKa = 11.84PLPTSQEE96 pKa = 4.33SFVSATDD103 pKa = 4.47DD104 pKa = 3.9IGLKK108 pKa = 10.7DD109 pKa = 4.57LFFEE113 pKa = 4.8PKK115 pKa = 8.91LTPEE119 pKa = 3.99TCSPYY124 pKa = 10.28QADD127 pKa = 3.42INIRR131 pKa = 11.84PLKK134 pKa = 10.33FFGPNGVLIKK144 pKa = 9.47RR145 pKa = 11.84TPVEE149 pKa = 3.83GSTPVTFPVTSEE161 pKa = 4.0NQSIPNNATITATFSEE177 pKa = 4.54DD178 pKa = 3.15RR179 pKa = 11.84QNLVIGGTGEE189 pKa = 4.76GIVQLRR195 pKa = 11.84FTWDD199 pKa = 3.58DD200 pKa = 3.53KK201 pKa = 11.52PSISGLAVGTLTVAGASFTQSGEE224 pKa = 4.01KK225 pKa = 10.81GSDD228 pKa = 3.28NDD230 pKa = 3.61STSIVTAGQSYY241 pKa = 9.44PITLSGNSGTSGSRR255 pKa = 11.84LQGNTQIEE263 pKa = 4.39YY264 pKa = 10.8DD265 pKa = 3.87DD266 pKa = 5.03DD267 pKa = 4.52VPGFDD272 pKa = 4.21VNATLSITEE281 pKa = 4.12VLPTTTSTNVAGYY294 pKa = 9.57WSDD297 pKa = 3.28EE298 pKa = 3.96GNKK301 pKa = 10.01YY302 pKa = 9.91GVWVNPAICTLPQQPQSVSYY322 pKa = 10.54KK323 pKa = 9.09ITIDD327 pKa = 3.42EE328 pKa = 4.48ADD330 pKa = 3.35TYY332 pKa = 11.46GFTFGCDD339 pKa = 3.2DD340 pKa = 4.35NATLTINDD348 pKa = 3.73EE349 pKa = 4.15TSAFLTAVGGIFEE362 pKa = 4.7GGSYY366 pKa = 7.55NTPYY370 pKa = 10.26TGTRR374 pKa = 11.84ALTAGTLILTVNCTNSDD391 pKa = 4.06AGFQDD396 pKa = 4.48ANGDD400 pKa = 4.03PTGLAYY406 pKa = 10.23DD407 pKa = 3.89WQRR410 pKa = 11.84NPGGWYY416 pKa = 9.74IKK418 pKa = 8.67MCKK421 pKa = 10.03GGLCASGTNINWVPVGPHH439 pKa = 6.26PAWSDD444 pKa = 3.58FMDD447 pKa = 4.81EE448 pKa = 4.32YY449 pKa = 11.67AVFPSNIDD457 pKa = 3.41PLLDD461 pKa = 3.32QTQQATWNINIPTTGNYY478 pKa = 8.38TFEE481 pKa = 4.57CSADD485 pKa = 3.48NNATFTLDD493 pKa = 3.08GTQIATSNSFTTTTSVSLTNISEE516 pKa = 4.57GPHH519 pKa = 5.19TLVVSVFNGTNSNGANEE536 pKa = 4.21WTTNPGGVAWRR547 pKa = 11.84IQYY550 pKa = 10.69GGGSIDD556 pKa = 5.95ANFQSNGRR564 pKa = 11.84LLVTGAGAARR574 pKa = 11.84LDD576 pKa = 5.42FDD578 pKa = 5.96FEE580 pKa = 4.43WDD582 pKa = 4.53DD583 pKa = 4.11NPSTYY588 pKa = 9.85DD589 pKa = 3.25TALGTVAYY597 pKa = 7.31PQLGVSFTQNTGSSSGSDD615 pKa = 2.92SDD617 pKa = 3.82NVNNVTAGDD626 pKa = 3.83YY627 pKa = 11.17GVTILNNSGGFTVEE641 pKa = 3.78NSGKK645 pKa = 9.95RR646 pKa = 11.84LCFKK650 pKa = 10.92DD651 pKa = 5.35LDD653 pKa = 4.46GNDD656 pKa = 4.12CNAQLDD662 pKa = 4.0LSIVQGDD669 pKa = 3.66ATIASSLDD677 pKa = 3.54LNTPGDD683 pKa = 4.26GNLFWHH689 pKa = 6.4TRR691 pKa = 11.84LATGYY696 pKa = 8.63TFLEE700 pKa = 4.92DD701 pKa = 3.27NN702 pKa = 4.34
Molecular weight: 75.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E3SIE5|E3SIE5_9CAUD Uncharacterized protein OS=Synechococcus phage S-SM1 OX=444859 GN=SSM1_138 PE=4 SV=1
MM1 pKa = 7.97RR2 pKa = 11.84GGKK5 pKa = 8.73MIPNSTYY12 pKa = 10.2IIVPSYY18 pKa = 10.56GVRR21 pKa = 11.84SCTIFPRR28 pKa = 11.84AINN31 pKa = 3.46
MM1 pKa = 7.97RR2 pKa = 11.84GGKK5 pKa = 8.73MIPNSTYY12 pKa = 10.2IIVPSYY18 pKa = 10.56GVRR21 pKa = 11.84SCTIFPRR28 pKa = 11.84AINN31 pKa = 3.46
Molecular weight: 3.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
56423 |
31 |
6180 |
241.1 |
26.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.203 ± 0.278 | 0.884 ± 0.115 |
6.692 ± 0.141 | 6.107 ± 0.317 |
4.333 ± 0.101 | 7.823 ± 0.399 |
1.581 ± 0.165 | 6.072 ± 0.185 |
5.718 ± 0.394 | 7.378 ± 0.205 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.191 ± 0.263 | 5.601 ± 0.186 |
3.97 ± 0.159 | 3.711 ± 0.097 |
4.13 ± 0.236 | 6.919 ± 0.225 |
7.669 ± 0.558 | 6.698 ± 0.236 |
1.102 ± 0.092 | 4.218 ± 0.134 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
