
Candidatus Saccharibacteria bacterium RAAC3_TM7_1
Taxonomy: cellular organisms; Bacteria; Bacteria incertae sedis; Bacteria candidate phyla; Candidatus Saccharibacteria; unclassified Saccharibacteria
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
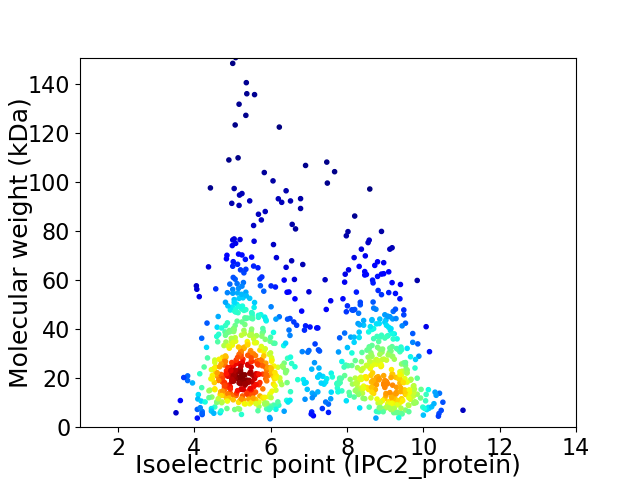
Virtual 2D-PAGE plot for 920 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V5RUT5|V5RUT5_9BACT Flavodoxin/nitric oxide synthase OS=Candidatus Saccharibacteria bacterium RAAC3_TM7_1 OX=1394711 GN=RAAC3_TM7C00001G0130 PE=4 SV=1
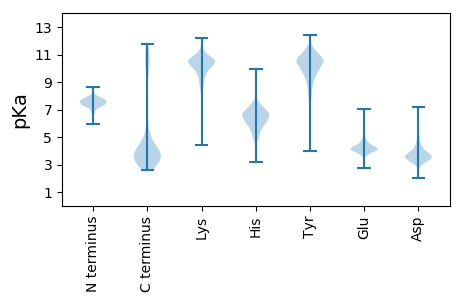
MM1 pKa = 7.42LSIRR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 7.85MLPGLSLILASLVFPIMFSTGAYY30 pKa = 9.66AVSQYY35 pKa = 11.68DD36 pKa = 3.23NAYY39 pKa = 10.43NHH41 pKa = 6.15VDD43 pKa = 3.23QLKK46 pKa = 10.0IVQNNYY52 pKa = 10.64NGVTCDD58 pKa = 3.86PLDD61 pKa = 3.15ITMTYY66 pKa = 10.48QSYY69 pKa = 11.18LNNSAKK75 pKa = 9.97WYY77 pKa = 6.12PTTSAFATTMQAYY90 pKa = 9.44KK91 pKa = 10.74SSFAQARR98 pKa = 11.84QAGSWTVSVVLVNDD112 pKa = 4.25GSGGYY117 pKa = 8.61TSIVRR122 pKa = 11.84INWFEE127 pKa = 4.48DD128 pKa = 3.74DD129 pKa = 3.66PGKK132 pKa = 10.74LVFKK136 pKa = 9.38GTSPNTQVEE145 pKa = 4.59YY146 pKa = 11.04QSTTDD151 pKa = 3.57TQGHH155 pKa = 5.47YY156 pKa = 11.11AFISTIDD163 pKa = 3.31FARR166 pKa = 11.84PYY168 pKa = 10.97GSTDD172 pKa = 3.07AGNCTPHH179 pKa = 7.4VYY181 pKa = 10.54AVGDD185 pKa = 4.1FVANANEE192 pKa = 3.7EE193 pKa = 4.42TYY195 pKa = 11.22AVVSTEE201 pKa = 4.68GAFSWTPTSSSEE213 pKa = 3.83MGHH216 pKa = 6.0WANLFATSTNVQYY229 pKa = 8.12PTGYY233 pKa = 9.89EE234 pKa = 3.99GAAISANSPTATYY247 pKa = 11.23VAMGDD252 pKa = 3.75SFSSGEE258 pKa = 3.97GNPPFEE264 pKa = 4.99YY265 pKa = 8.98GTDD268 pKa = 3.55SASDD272 pKa = 3.31KK273 pKa = 10.68CHH275 pKa = 6.98RR276 pKa = 11.84SPNAYY281 pKa = 8.87PRR283 pKa = 11.84LLQSDD288 pKa = 4.02SSLSLGSTAFVACSGATTDD307 pKa = 4.73DD308 pKa = 3.7VLNGGSGAGNWGEE321 pKa = 4.19GPQVDD326 pKa = 4.48ALLSDD331 pKa = 4.08TEE333 pKa = 4.59VVTITIGGNNIKK345 pKa = 10.21FAEE348 pKa = 4.42FAHH351 pKa = 6.26ACLFGSCNSSSDD363 pKa = 3.8EE364 pKa = 4.11YY365 pKa = 11.01QEE367 pKa = 4.32SWDD370 pKa = 3.63VMTDD374 pKa = 3.35LTRR377 pKa = 11.84SDD379 pKa = 4.27YY380 pKa = 11.27LPSQLDD386 pKa = 3.57SLFSDD391 pKa = 3.72IAFHH395 pKa = 6.72LWQNTSVKK403 pKa = 9.78VYY405 pKa = 9.96VVGYY409 pKa = 8.71PYY411 pKa = 10.96VITQASWNDD420 pKa = 3.43RR421 pKa = 11.84GVGICSDD428 pKa = 3.77FDD430 pKa = 3.82EE431 pKa = 6.41DD432 pKa = 3.68EE433 pKa = 5.37AIAAEE438 pKa = 4.47TIVSKK443 pKa = 10.97LDD445 pKa = 3.55TVIEE449 pKa = 4.16TAVTDD454 pKa = 3.96FDD456 pKa = 4.33DD457 pKa = 3.97SRR459 pKa = 11.84FVYY462 pKa = 10.41VDD464 pKa = 3.55PLATGSPFLGHH475 pKa = 5.81EE476 pKa = 4.31LCRR479 pKa = 11.84NGSYY483 pKa = 10.93FNGIEE488 pKa = 4.0TSGALGGDD496 pKa = 3.81DD497 pKa = 4.91AYY499 pKa = 11.65VFHH502 pKa = 8.09PDD504 pKa = 3.13TNGQQAYY511 pKa = 10.19ADD513 pKa = 5.12LIRR516 pKa = 11.84SYY518 pKa = 10.8MSS520 pKa = 3.07
MM1 pKa = 7.42LSIRR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 7.85MLPGLSLILASLVFPIMFSTGAYY30 pKa = 9.66AVSQYY35 pKa = 11.68DD36 pKa = 3.23NAYY39 pKa = 10.43NHH41 pKa = 6.15VDD43 pKa = 3.23QLKK46 pKa = 10.0IVQNNYY52 pKa = 10.64NGVTCDD58 pKa = 3.86PLDD61 pKa = 3.15ITMTYY66 pKa = 10.48QSYY69 pKa = 11.18LNNSAKK75 pKa = 9.97WYY77 pKa = 6.12PTTSAFATTMQAYY90 pKa = 9.44KK91 pKa = 10.74SSFAQARR98 pKa = 11.84QAGSWTVSVVLVNDD112 pKa = 4.25GSGGYY117 pKa = 8.61TSIVRR122 pKa = 11.84INWFEE127 pKa = 4.48DD128 pKa = 3.74DD129 pKa = 3.66PGKK132 pKa = 10.74LVFKK136 pKa = 9.38GTSPNTQVEE145 pKa = 4.59YY146 pKa = 11.04QSTTDD151 pKa = 3.57TQGHH155 pKa = 5.47YY156 pKa = 11.11AFISTIDD163 pKa = 3.31FARR166 pKa = 11.84PYY168 pKa = 10.97GSTDD172 pKa = 3.07AGNCTPHH179 pKa = 7.4VYY181 pKa = 10.54AVGDD185 pKa = 4.1FVANANEE192 pKa = 3.7EE193 pKa = 4.42TYY195 pKa = 11.22AVVSTEE201 pKa = 4.68GAFSWTPTSSSEE213 pKa = 3.83MGHH216 pKa = 6.0WANLFATSTNVQYY229 pKa = 8.12PTGYY233 pKa = 9.89EE234 pKa = 3.99GAAISANSPTATYY247 pKa = 11.23VAMGDD252 pKa = 3.75SFSSGEE258 pKa = 3.97GNPPFEE264 pKa = 4.99YY265 pKa = 8.98GTDD268 pKa = 3.55SASDD272 pKa = 3.31KK273 pKa = 10.68CHH275 pKa = 6.98RR276 pKa = 11.84SPNAYY281 pKa = 8.87PRR283 pKa = 11.84LLQSDD288 pKa = 4.02SSLSLGSTAFVACSGATTDD307 pKa = 4.73DD308 pKa = 3.7VLNGGSGAGNWGEE321 pKa = 4.19GPQVDD326 pKa = 4.48ALLSDD331 pKa = 4.08TEE333 pKa = 4.59VVTITIGGNNIKK345 pKa = 10.21FAEE348 pKa = 4.42FAHH351 pKa = 6.26ACLFGSCNSSSDD363 pKa = 3.8EE364 pKa = 4.11YY365 pKa = 11.01QEE367 pKa = 4.32SWDD370 pKa = 3.63VMTDD374 pKa = 3.35LTRR377 pKa = 11.84SDD379 pKa = 4.27YY380 pKa = 11.27LPSQLDD386 pKa = 3.57SLFSDD391 pKa = 3.72IAFHH395 pKa = 6.72LWQNTSVKK403 pKa = 9.78VYY405 pKa = 9.96VVGYY409 pKa = 8.71PYY411 pKa = 10.96VITQASWNDD420 pKa = 3.43RR421 pKa = 11.84GVGICSDD428 pKa = 3.77FDD430 pKa = 3.82EE431 pKa = 6.41DD432 pKa = 3.68EE433 pKa = 5.37AIAAEE438 pKa = 4.47TIVSKK443 pKa = 10.97LDD445 pKa = 3.55TVIEE449 pKa = 4.16TAVTDD454 pKa = 3.96FDD456 pKa = 4.33DD457 pKa = 3.97SRR459 pKa = 11.84FVYY462 pKa = 10.41VDD464 pKa = 3.55PLATGSPFLGHH475 pKa = 5.81EE476 pKa = 4.31LCRR479 pKa = 11.84NGSYY483 pKa = 10.93FNGIEE488 pKa = 4.0TSGALGGDD496 pKa = 3.81DD497 pKa = 4.91AYY499 pKa = 11.65VFHH502 pKa = 8.09PDD504 pKa = 3.13TNGQQAYY511 pKa = 10.19ADD513 pKa = 5.12LIRR516 pKa = 11.84SYY518 pKa = 10.8MSS520 pKa = 3.07
Molecular weight: 56.19 kDa
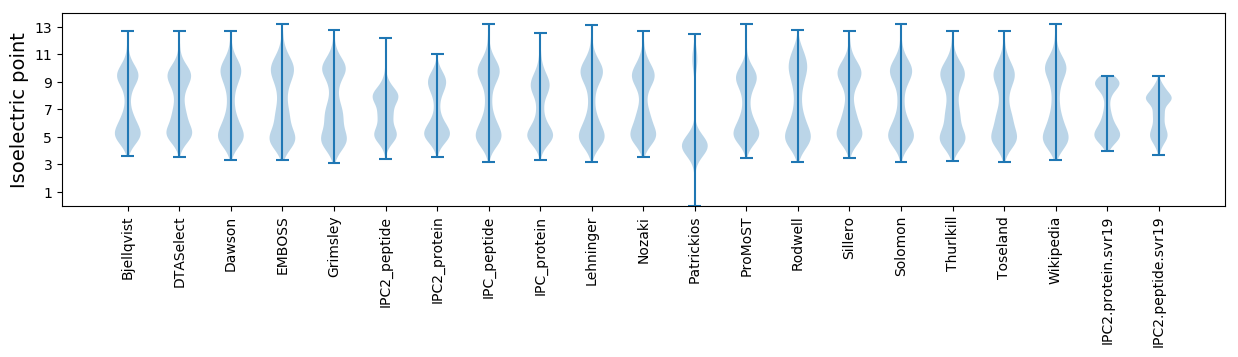
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V5RVU6|V5RVU6_9BACT Fimbrial protein pilin OS=Candidatus Saccharibacteria bacterium RAAC3_TM7_1 OX=1394711 GN=RAAC3_TM7C00001G0659 PE=4 SV=1
MM1 pKa = 7.84PKK3 pKa = 10.55LKK5 pKa = 8.66THH7 pKa = 6.85KK8 pKa = 9.66GTAKK12 pKa = 9.88RR13 pKa = 11.84VKK15 pKa = 8.75ITGSGKK21 pKa = 8.17ITRR24 pKa = 11.84QRR26 pKa = 11.84ASGGHH31 pKa = 6.27LLAKK35 pKa = 9.92KK36 pKa = 10.13SKK38 pKa = 9.24SRR40 pKa = 11.84KK41 pKa = 8.36RR42 pKa = 11.84AINSTGAITGRR53 pKa = 11.84IAKK56 pKa = 9.64NIKK59 pKa = 9.07QALGAKK65 pKa = 9.35
MM1 pKa = 7.84PKK3 pKa = 10.55LKK5 pKa = 8.66THH7 pKa = 6.85KK8 pKa = 9.66GTAKK12 pKa = 9.88RR13 pKa = 11.84VKK15 pKa = 8.75ITGSGKK21 pKa = 8.17ITRR24 pKa = 11.84QRR26 pKa = 11.84ASGGHH31 pKa = 6.27LLAKK35 pKa = 9.92KK36 pKa = 10.13SKK38 pKa = 9.24SRR40 pKa = 11.84KK41 pKa = 8.36RR42 pKa = 11.84AINSTGAITGRR53 pKa = 11.84IAKK56 pKa = 9.64NIKK59 pKa = 9.07QALGAKK65 pKa = 9.35
Molecular weight: 6.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
255811 |
30 |
1341 |
278.1 |
30.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.995 ± 0.089 | 0.602 ± 0.028 |
5.698 ± 0.072 | 6.273 ± 0.097 |
3.647 ± 0.058 | 7.011 ± 0.072 |
2.112 ± 0.046 | 6.468 ± 0.059 |
5.924 ± 0.088 | 9.67 ± 0.096 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.27 ± 0.036 | 3.717 ± 0.06 |
3.963 ± 0.052 | 3.736 ± 0.047 |
5.47 ± 0.079 | 6.327 ± 0.08 |
6.216 ± 0.081 | 7.329 ± 0.072 |
1.112 ± 0.035 | 3.458 ± 0.055 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
