
Miniopterus bat coronavirus HKU8
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Minunacovirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
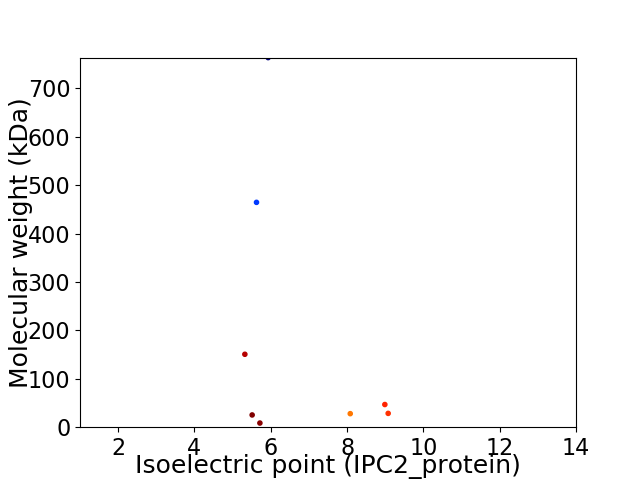
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
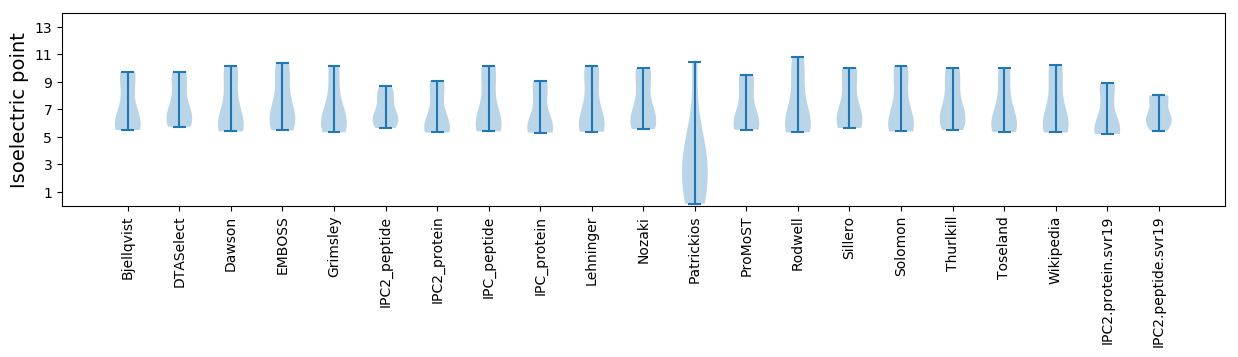
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B1PHK3|B1PHK3_9ALPC ORF3 protein OS=Miniopterus bat coronavirus HKU8 OX=694001 GN=ORF3 PE=4 SV=1
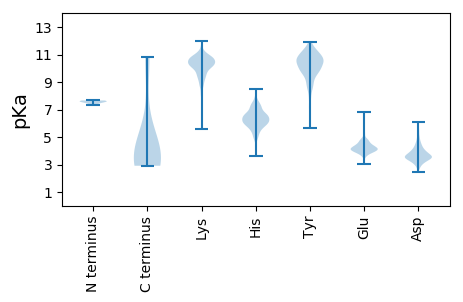
MM1 pKa = 7.66KK2 pKa = 10.42SLLVLSLLALLATLSVNAQVTGEE25 pKa = 4.15GGVAEE30 pKa = 4.18GNYY33 pKa = 8.3WVNASCAGWSYY44 pKa = 10.71FYY46 pKa = 11.12ALKK49 pKa = 10.83LGLPPNASAIVTGYY63 pKa = 10.35LPKK66 pKa = 10.29PKK68 pKa = 10.24GWICPRR74 pKa = 11.84FNGAGIYY81 pKa = 8.3TRR83 pKa = 11.84NNANAVFVMYY93 pKa = 8.47RR94 pKa = 11.84TKK96 pKa = 11.11ALAFEE101 pKa = 4.84IGVSSSAGGEE111 pKa = 3.98QYY113 pKa = 10.6SAYY116 pKa = 9.63MAQQNGKK123 pKa = 8.88YY124 pKa = 9.79LVLRR128 pKa = 11.84ICKK131 pKa = 7.98WQNGTLAAPTLQSTSGKK148 pKa = 10.31DD149 pKa = 3.13CIVNVKK155 pKa = 9.65VDD157 pKa = 3.07NHH159 pKa = 5.27MFYY162 pKa = 10.53HH163 pKa = 6.83AAHH166 pKa = 7.47DD167 pKa = 3.76IVGMSWSGDD176 pKa = 3.07AVRR179 pKa = 11.84LYY181 pKa = 9.94TQTDD185 pKa = 3.34TKK187 pKa = 10.59TYY189 pKa = 9.62YY190 pKa = 9.98IPNSWDD196 pKa = 3.26RR197 pKa = 11.84VSIRR201 pKa = 11.84CPDD204 pKa = 3.37KK205 pKa = 10.83FSCSSQIVTKK215 pKa = 10.64AITVNVTTFANGTIDD230 pKa = 5.16KK231 pKa = 10.86YY232 pKa = 11.26AICDD236 pKa = 3.47NCNGYY241 pKa = 8.34PAHH244 pKa = 7.13IFPVSEE250 pKa = 4.43GGLIPADD257 pKa = 3.76FNFSNWFLLTNSSTIVDD274 pKa = 3.49GRR276 pKa = 11.84IVSEE280 pKa = 4.14QPVLLMCLWAVPGLMSTNSFVYY302 pKa = 10.29FNGTAPNKK310 pKa = 9.21QCNGYY315 pKa = 8.38ATDD318 pKa = 3.68SAFEE322 pKa = 3.95ALRR325 pKa = 11.84FSLNFTDD332 pKa = 3.56EE333 pKa = 4.2RR334 pKa = 11.84VFAGSGSVVLLVSGLQYY351 pKa = 11.15KK352 pKa = 8.68FSCTNNSEE360 pKa = 4.04AVIDD364 pKa = 3.77SGIPFGNVVEE374 pKa = 4.3PFYY377 pKa = 11.22CFVSINGTSIFVGMLPAVLRR397 pKa = 11.84EE398 pKa = 4.09IVITRR403 pKa = 11.84YY404 pKa = 10.47GSIYY408 pKa = 10.64LNGFSIFQGPPIQGVLFNVTNRR430 pKa = 11.84GATDD434 pKa = 3.54LWTVALSNFTEE445 pKa = 4.36VLAEE449 pKa = 4.15VQSTAIKK456 pKa = 10.57ALLYY460 pKa = 10.79CDD462 pKa = 5.71DD463 pKa = 5.18PLSQLKK469 pKa = 9.76CQQLQFSLPDD479 pKa = 3.36GFYY482 pKa = 9.99ATASLFQHH490 pKa = 6.22EE491 pKa = 4.7LPRR494 pKa = 11.84TFVTLPRR501 pKa = 11.84HH502 pKa = 5.43FTHH505 pKa = 6.23SWINLRR511 pKa = 11.84IKK513 pKa = 9.01WKK515 pKa = 10.89NGVCYY520 pKa = 9.86NCPPASSWIDD530 pKa = 3.05FVTFNSNGTEE540 pKa = 3.83NVLPEE545 pKa = 3.85RR546 pKa = 11.84TLCVNTTQFTTNLTLIEE563 pKa = 4.21EE564 pKa = 4.58AFSYY568 pKa = 7.75STPVVVRR575 pKa = 11.84ADD577 pKa = 3.48DD578 pKa = 3.99CPFDD582 pKa = 4.04FQSLNNYY589 pKa = 7.66LTFGSICFSLNGTIGKK605 pKa = 9.74GCTLGIYY612 pKa = 9.81KK613 pKa = 10.15RR614 pKa = 11.84ASSQYY619 pKa = 9.3IPIWNVWVAYY629 pKa = 9.37TSGDD633 pKa = 3.44NILGVRR639 pKa = 11.84EE640 pKa = 4.12PNVGVRR646 pKa = 11.84DD647 pKa = 3.71QSVVHH652 pKa = 5.84QNVCTSYY659 pKa = 10.77TIFGHH664 pKa = 5.19SGRR667 pKa = 11.84GIIRR671 pKa = 11.84PANISYY677 pKa = 10.41IAGVYY682 pKa = 7.3YY683 pKa = 9.93TAASGQLLGFKK694 pKa = 9.21NTTTGEE700 pKa = 4.34VFSVTPCNPSQQAVVVKK717 pKa = 10.44DD718 pKa = 3.52RR719 pKa = 11.84LVGVMSSTSTVSIPFNNTIPTPSFYY744 pKa = 10.91YY745 pKa = 10.32HH746 pKa = 6.95SNATSSCDD754 pKa = 3.6DD755 pKa = 3.81PSVVYY760 pKa = 10.1SSIGICDD767 pKa = 3.68DD768 pKa = 3.86GGITFVNSTRR778 pKa = 11.84VRR780 pKa = 11.84GEE782 pKa = 3.65PDD784 pKa = 3.08PAISMGNISVPSNFTVSIQVEE805 pKa = 4.26YY806 pKa = 10.4LQMSIRR812 pKa = 11.84PVSIDD817 pKa = 2.82CAMYY821 pKa = 9.32VCNGNPHH828 pKa = 5.96CTRR831 pKa = 11.84LLQQYY836 pKa = 10.04ISACRR841 pKa = 11.84TIEE844 pKa = 3.89EE845 pKa = 4.26ALQLSARR852 pKa = 11.84LEE854 pKa = 4.25SFEE857 pKa = 4.42VNSMLTVSEE866 pKa = 4.24TALDD870 pKa = 3.76LVNISTFGGDD880 pKa = 3.66YY881 pKa = 10.92NLTALLPQGGGKK893 pKa = 9.86RR894 pKa = 11.84SVIEE898 pKa = 5.43DD899 pKa = 3.28ILFDD903 pKa = 3.79KK904 pKa = 11.1VVTSGLGTVDD914 pKa = 3.79EE915 pKa = 4.9DD916 pKa = 4.87YY917 pKa = 11.2KK918 pKa = 11.11RR919 pKa = 11.84CTNGIGIADD928 pKa = 4.13VPCAQYY934 pKa = 11.47YY935 pKa = 9.06NGIMVLPGVVDD946 pKa = 3.75EE947 pKa = 5.2EE948 pKa = 4.58KK949 pKa = 10.47MSIYY953 pKa = 9.62TASLLGGMTMGGFTPVAALPFALSVQSRR981 pKa = 11.84LNYY984 pKa = 9.76VALQTDD990 pKa = 4.37VLQKK994 pKa = 10.47NQQILANAFNSAIGNITVAFDD1015 pKa = 3.48QVTTAVQQTSDD1026 pKa = 3.63AIKK1029 pKa = 9.55TVASALNKK1037 pKa = 8.89VQSVVNSQGQALHH1050 pKa = 6.21QLTKK1054 pKa = 10.62QLASNFQAISASIEE1068 pKa = 4.23DD1069 pKa = 3.6IYY1071 pKa = 11.82NRR1073 pKa = 11.84LDD1075 pKa = 3.61GLAADD1080 pKa = 4.45ANVDD1084 pKa = 3.63RR1085 pKa = 11.84LITGRR1090 pKa = 11.84LAALNAFVTQTLTKK1104 pKa = 8.05YY1105 pKa = 9.0TEE1107 pKa = 4.09VRR1109 pKa = 11.84ASRR1112 pKa = 11.84LLAQEE1117 pKa = 5.01KK1118 pKa = 10.43INEE1121 pKa = 4.44CVKK1124 pKa = 10.72SQSTRR1129 pKa = 11.84YY1130 pKa = 8.87GFCGNGTHH1138 pKa = 7.47LFSIPNAAPEE1148 pKa = 4.81GIMLFHH1154 pKa = 6.48TVLVPTEE1161 pKa = 4.22YY1162 pKa = 11.1VSVTAWSGYY1171 pKa = 6.77CHH1173 pKa = 6.83NGVGYY1178 pKa = 10.37AVKK1181 pKa = 10.57DD1182 pKa = 3.52VGNSLFQFNNTFYY1195 pKa = 9.59ITPRR1199 pKa = 11.84NMYY1202 pKa = 9.48QPRR1205 pKa = 11.84TPTSADD1211 pKa = 4.29FIRR1214 pKa = 11.84ISGCNVVYY1222 pKa = 10.88VNITDD1227 pKa = 3.56EE1228 pKa = 4.3QLPQVQPEE1236 pKa = 4.49FIDD1239 pKa = 3.57VNKK1242 pKa = 9.54TLEE1245 pKa = 4.2EE1246 pKa = 4.0LMSRR1250 pKa = 11.84LPNNTGPNLPLDD1262 pKa = 4.22IFNQTYY1268 pKa = 10.88LNISAEE1274 pKa = 3.89IDD1276 pKa = 3.29ALEE1279 pKa = 4.32NKK1281 pKa = 10.14SLEE1284 pKa = 4.33LQATADD1290 pKa = 3.78KK1291 pKa = 10.79LQLTIEE1297 pKa = 4.19QLNATLVDD1305 pKa = 4.35LEE1307 pKa = 4.14WLNRR1311 pKa = 11.84FEE1313 pKa = 6.27QYY1315 pKa = 10.86VKK1317 pKa = 9.92WPWWVWLTMIIALVLLTGLMLWCCLATGCCGCCSCMASTLDD1358 pKa = 3.63FRR1360 pKa = 11.84GSRR1363 pKa = 11.84LQQYY1367 pKa = 7.6EE1368 pKa = 4.15VEE1370 pKa = 4.46KK1371 pKa = 11.13VHH1373 pKa = 6.56IQQ1375 pKa = 3.09
MM1 pKa = 7.66KK2 pKa = 10.42SLLVLSLLALLATLSVNAQVTGEE25 pKa = 4.15GGVAEE30 pKa = 4.18GNYY33 pKa = 8.3WVNASCAGWSYY44 pKa = 10.71FYY46 pKa = 11.12ALKK49 pKa = 10.83LGLPPNASAIVTGYY63 pKa = 10.35LPKK66 pKa = 10.29PKK68 pKa = 10.24GWICPRR74 pKa = 11.84FNGAGIYY81 pKa = 8.3TRR83 pKa = 11.84NNANAVFVMYY93 pKa = 8.47RR94 pKa = 11.84TKK96 pKa = 11.11ALAFEE101 pKa = 4.84IGVSSSAGGEE111 pKa = 3.98QYY113 pKa = 10.6SAYY116 pKa = 9.63MAQQNGKK123 pKa = 8.88YY124 pKa = 9.79LVLRR128 pKa = 11.84ICKK131 pKa = 7.98WQNGTLAAPTLQSTSGKK148 pKa = 10.31DD149 pKa = 3.13CIVNVKK155 pKa = 9.65VDD157 pKa = 3.07NHH159 pKa = 5.27MFYY162 pKa = 10.53HH163 pKa = 6.83AAHH166 pKa = 7.47DD167 pKa = 3.76IVGMSWSGDD176 pKa = 3.07AVRR179 pKa = 11.84LYY181 pKa = 9.94TQTDD185 pKa = 3.34TKK187 pKa = 10.59TYY189 pKa = 9.62YY190 pKa = 9.98IPNSWDD196 pKa = 3.26RR197 pKa = 11.84VSIRR201 pKa = 11.84CPDD204 pKa = 3.37KK205 pKa = 10.83FSCSSQIVTKK215 pKa = 10.64AITVNVTTFANGTIDD230 pKa = 5.16KK231 pKa = 10.86YY232 pKa = 11.26AICDD236 pKa = 3.47NCNGYY241 pKa = 8.34PAHH244 pKa = 7.13IFPVSEE250 pKa = 4.43GGLIPADD257 pKa = 3.76FNFSNWFLLTNSSTIVDD274 pKa = 3.49GRR276 pKa = 11.84IVSEE280 pKa = 4.14QPVLLMCLWAVPGLMSTNSFVYY302 pKa = 10.29FNGTAPNKK310 pKa = 9.21QCNGYY315 pKa = 8.38ATDD318 pKa = 3.68SAFEE322 pKa = 3.95ALRR325 pKa = 11.84FSLNFTDD332 pKa = 3.56EE333 pKa = 4.2RR334 pKa = 11.84VFAGSGSVVLLVSGLQYY351 pKa = 11.15KK352 pKa = 8.68FSCTNNSEE360 pKa = 4.04AVIDD364 pKa = 3.77SGIPFGNVVEE374 pKa = 4.3PFYY377 pKa = 11.22CFVSINGTSIFVGMLPAVLRR397 pKa = 11.84EE398 pKa = 4.09IVITRR403 pKa = 11.84YY404 pKa = 10.47GSIYY408 pKa = 10.64LNGFSIFQGPPIQGVLFNVTNRR430 pKa = 11.84GATDD434 pKa = 3.54LWTVALSNFTEE445 pKa = 4.36VLAEE449 pKa = 4.15VQSTAIKK456 pKa = 10.57ALLYY460 pKa = 10.79CDD462 pKa = 5.71DD463 pKa = 5.18PLSQLKK469 pKa = 9.76CQQLQFSLPDD479 pKa = 3.36GFYY482 pKa = 9.99ATASLFQHH490 pKa = 6.22EE491 pKa = 4.7LPRR494 pKa = 11.84TFVTLPRR501 pKa = 11.84HH502 pKa = 5.43FTHH505 pKa = 6.23SWINLRR511 pKa = 11.84IKK513 pKa = 9.01WKK515 pKa = 10.89NGVCYY520 pKa = 9.86NCPPASSWIDD530 pKa = 3.05FVTFNSNGTEE540 pKa = 3.83NVLPEE545 pKa = 3.85RR546 pKa = 11.84TLCVNTTQFTTNLTLIEE563 pKa = 4.21EE564 pKa = 4.58AFSYY568 pKa = 7.75STPVVVRR575 pKa = 11.84ADD577 pKa = 3.48DD578 pKa = 3.99CPFDD582 pKa = 4.04FQSLNNYY589 pKa = 7.66LTFGSICFSLNGTIGKK605 pKa = 9.74GCTLGIYY612 pKa = 9.81KK613 pKa = 10.15RR614 pKa = 11.84ASSQYY619 pKa = 9.3IPIWNVWVAYY629 pKa = 9.37TSGDD633 pKa = 3.44NILGVRR639 pKa = 11.84EE640 pKa = 4.12PNVGVRR646 pKa = 11.84DD647 pKa = 3.71QSVVHH652 pKa = 5.84QNVCTSYY659 pKa = 10.77TIFGHH664 pKa = 5.19SGRR667 pKa = 11.84GIIRR671 pKa = 11.84PANISYY677 pKa = 10.41IAGVYY682 pKa = 7.3YY683 pKa = 9.93TAASGQLLGFKK694 pKa = 9.21NTTTGEE700 pKa = 4.34VFSVTPCNPSQQAVVVKK717 pKa = 10.44DD718 pKa = 3.52RR719 pKa = 11.84LVGVMSSTSTVSIPFNNTIPTPSFYY744 pKa = 10.91YY745 pKa = 10.32HH746 pKa = 6.95SNATSSCDD754 pKa = 3.6DD755 pKa = 3.81PSVVYY760 pKa = 10.1SSIGICDD767 pKa = 3.68DD768 pKa = 3.86GGITFVNSTRR778 pKa = 11.84VRR780 pKa = 11.84GEE782 pKa = 3.65PDD784 pKa = 3.08PAISMGNISVPSNFTVSIQVEE805 pKa = 4.26YY806 pKa = 10.4LQMSIRR812 pKa = 11.84PVSIDD817 pKa = 2.82CAMYY821 pKa = 9.32VCNGNPHH828 pKa = 5.96CTRR831 pKa = 11.84LLQQYY836 pKa = 10.04ISACRR841 pKa = 11.84TIEE844 pKa = 3.89EE845 pKa = 4.26ALQLSARR852 pKa = 11.84LEE854 pKa = 4.25SFEE857 pKa = 4.42VNSMLTVSEE866 pKa = 4.24TALDD870 pKa = 3.76LVNISTFGGDD880 pKa = 3.66YY881 pKa = 10.92NLTALLPQGGGKK893 pKa = 9.86RR894 pKa = 11.84SVIEE898 pKa = 5.43DD899 pKa = 3.28ILFDD903 pKa = 3.79KK904 pKa = 11.1VVTSGLGTVDD914 pKa = 3.79EE915 pKa = 4.9DD916 pKa = 4.87YY917 pKa = 11.2KK918 pKa = 11.11RR919 pKa = 11.84CTNGIGIADD928 pKa = 4.13VPCAQYY934 pKa = 11.47YY935 pKa = 9.06NGIMVLPGVVDD946 pKa = 3.75EE947 pKa = 5.2EE948 pKa = 4.58KK949 pKa = 10.47MSIYY953 pKa = 9.62TASLLGGMTMGGFTPVAALPFALSVQSRR981 pKa = 11.84LNYY984 pKa = 9.76VALQTDD990 pKa = 4.37VLQKK994 pKa = 10.47NQQILANAFNSAIGNITVAFDD1015 pKa = 3.48QVTTAVQQTSDD1026 pKa = 3.63AIKK1029 pKa = 9.55TVASALNKK1037 pKa = 8.89VQSVVNSQGQALHH1050 pKa = 6.21QLTKK1054 pKa = 10.62QLASNFQAISASIEE1068 pKa = 4.23DD1069 pKa = 3.6IYY1071 pKa = 11.82NRR1073 pKa = 11.84LDD1075 pKa = 3.61GLAADD1080 pKa = 4.45ANVDD1084 pKa = 3.63RR1085 pKa = 11.84LITGRR1090 pKa = 11.84LAALNAFVTQTLTKK1104 pKa = 8.05YY1105 pKa = 9.0TEE1107 pKa = 4.09VRR1109 pKa = 11.84ASRR1112 pKa = 11.84LLAQEE1117 pKa = 5.01KK1118 pKa = 10.43INEE1121 pKa = 4.44CVKK1124 pKa = 10.72SQSTRR1129 pKa = 11.84YY1130 pKa = 8.87GFCGNGTHH1138 pKa = 7.47LFSIPNAAPEE1148 pKa = 4.81GIMLFHH1154 pKa = 6.48TVLVPTEE1161 pKa = 4.22YY1162 pKa = 11.1VSVTAWSGYY1171 pKa = 6.77CHH1173 pKa = 6.83NGVGYY1178 pKa = 10.37AVKK1181 pKa = 10.57DD1182 pKa = 3.52VGNSLFQFNNTFYY1195 pKa = 9.59ITPRR1199 pKa = 11.84NMYY1202 pKa = 9.48QPRR1205 pKa = 11.84TPTSADD1211 pKa = 4.29FIRR1214 pKa = 11.84ISGCNVVYY1222 pKa = 10.88VNITDD1227 pKa = 3.56EE1228 pKa = 4.3QLPQVQPEE1236 pKa = 4.49FIDD1239 pKa = 3.57VNKK1242 pKa = 9.54TLEE1245 pKa = 4.2EE1246 pKa = 4.0LMSRR1250 pKa = 11.84LPNNTGPNLPLDD1262 pKa = 4.22IFNQTYY1268 pKa = 10.88LNISAEE1274 pKa = 3.89IDD1276 pKa = 3.29ALEE1279 pKa = 4.32NKK1281 pKa = 10.14SLEE1284 pKa = 4.33LQATADD1290 pKa = 3.78KK1291 pKa = 10.79LQLTIEE1297 pKa = 4.19QLNATLVDD1305 pKa = 4.35LEE1307 pKa = 4.14WLNRR1311 pKa = 11.84FEE1313 pKa = 6.27QYY1315 pKa = 10.86VKK1317 pKa = 9.92WPWWVWLTMIIALVLLTGLMLWCCLATGCCGCCSCMASTLDD1358 pKa = 3.63FRR1360 pKa = 11.84GSRR1363 pKa = 11.84LQQYY1367 pKa = 7.6EE1368 pKa = 4.15VEE1370 pKa = 4.46KK1371 pKa = 11.13VHH1373 pKa = 6.56IQQ1375 pKa = 3.09
Molecular weight: 150.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0Q099|Q0Q099_9ALPC Nucleoprotein OS=Miniopterus bat coronavirus HKU8 OX=694001 GN=N PE=4 SV=1
MM1 pKa = 7.65FLRR4 pKa = 11.84KK5 pKa = 7.87TRR7 pKa = 11.84TSKK10 pKa = 10.89SSLIRR15 pKa = 11.84LMLIWILLVRR25 pKa = 11.84KK26 pKa = 7.87NQKK29 pKa = 10.19SSVNPRR35 pKa = 11.84KK36 pKa = 9.69RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.06KK41 pKa = 9.85HH42 pKa = 5.79LLNSTLQHH50 pKa = 6.43LCLHH54 pKa = 6.41HH55 pKa = 6.38QLCCQMLLPMLSLTWWMRR73 pKa = 11.84LLMPIMSLLHH83 pKa = 5.94EE84 pKa = 4.94LSTNKK89 pKa = 9.2MFVLALFAVLAIVNSAPIVNHH110 pKa = 6.35KK111 pKa = 9.91VLNPYY116 pKa = 7.64DD117 pKa = 3.97TPHH120 pKa = 6.51SGFRR124 pKa = 11.84GSCEE128 pKa = 3.71QLALTCRR135 pKa = 11.84LNVQPDD141 pKa = 3.66AFHH144 pKa = 7.11AAATTGSPVQIPRR157 pKa = 11.84ACVSFRR163 pKa = 11.84TVCSAWPKK171 pKa = 10.41YY172 pKa = 10.62DD173 pKa = 3.48FGYY176 pKa = 11.15LMSPDD181 pKa = 3.72VYY183 pKa = 11.28VVTTTPDD190 pKa = 2.67GRR192 pKa = 11.84YY193 pKa = 9.5KK194 pKa = 11.11YY195 pKa = 9.58EE196 pKa = 4.25LNSLAFTPEE205 pKa = 3.87GAQLFYY211 pKa = 10.99TLVDD215 pKa = 4.66GYY217 pKa = 11.22AHH219 pKa = 6.81EE220 pKa = 4.15RR221 pKa = 11.84RR222 pKa = 11.84KK223 pKa = 10.32LIKK226 pKa = 9.59EE227 pKa = 3.95GRR229 pKa = 11.84RR230 pKa = 11.84EE231 pKa = 3.9EE232 pKa = 4.6LPNLDD237 pKa = 3.82AEE239 pKa = 4.78YY240 pKa = 9.81IIHH243 pKa = 6.16PTEE246 pKa = 4.08SQQ248 pKa = 2.91
MM1 pKa = 7.65FLRR4 pKa = 11.84KK5 pKa = 7.87TRR7 pKa = 11.84TSKK10 pKa = 10.89SSLIRR15 pKa = 11.84LMLIWILLVRR25 pKa = 11.84KK26 pKa = 7.87NQKK29 pKa = 10.19SSVNPRR35 pKa = 11.84KK36 pKa = 9.69RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.06KK41 pKa = 9.85HH42 pKa = 5.79LLNSTLQHH50 pKa = 6.43LCLHH54 pKa = 6.41HH55 pKa = 6.38QLCCQMLLPMLSLTWWMRR73 pKa = 11.84LLMPIMSLLHH83 pKa = 5.94EE84 pKa = 4.94LSTNKK89 pKa = 9.2MFVLALFAVLAIVNSAPIVNHH110 pKa = 6.35KK111 pKa = 9.91VLNPYY116 pKa = 7.64DD117 pKa = 3.97TPHH120 pKa = 6.51SGFRR124 pKa = 11.84GSCEE128 pKa = 3.71QLALTCRR135 pKa = 11.84LNVQPDD141 pKa = 3.66AFHH144 pKa = 7.11AAATTGSPVQIPRR157 pKa = 11.84ACVSFRR163 pKa = 11.84TVCSAWPKK171 pKa = 10.41YY172 pKa = 10.62DD173 pKa = 3.48FGYY176 pKa = 11.15LMSPDD181 pKa = 3.72VYY183 pKa = 11.28VVTTTPDD190 pKa = 2.67GRR192 pKa = 11.84YY193 pKa = 9.5KK194 pKa = 11.11YY195 pKa = 9.58EE196 pKa = 4.25LNSLAFTPEE205 pKa = 3.87GAQLFYY211 pKa = 10.99TLVDD215 pKa = 4.66GYY217 pKa = 11.22AHH219 pKa = 6.81EE220 pKa = 4.15RR221 pKa = 11.84RR222 pKa = 11.84KK223 pKa = 10.32LIKK226 pKa = 9.59EE227 pKa = 3.95GRR229 pKa = 11.84RR230 pKa = 11.84EE231 pKa = 3.9EE232 pKa = 4.6LPNLDD237 pKa = 3.82AEE239 pKa = 4.78YY240 pKa = 9.81IIHH243 pKa = 6.16PTEE246 pKa = 4.08SQQ248 pKa = 2.91
Molecular weight: 28.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13719 |
74 |
6896 |
1714.9 |
189.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.508 ± 0.238 | 3.419 ± 0.368 |
5.299 ± 0.375 | 4.082 ± 0.202 |
5.882 ± 0.305 | 6.939 ± 0.354 |
1.793 ± 0.19 | 4.578 ± 0.454 |
5.255 ± 0.521 | 8.317 ± 0.586 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.012 ± 0.114 | 5.285 ± 0.312 |
4.002 ± 0.46 | 2.784 ± 0.612 |
3.564 ± 0.361 | 6.874 ± 0.483 |
6.218 ± 0.319 | 10.628 ± 0.711 |
1.225 ± 0.132 | 4.337 ± 0.311 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
