
Beihai toti-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
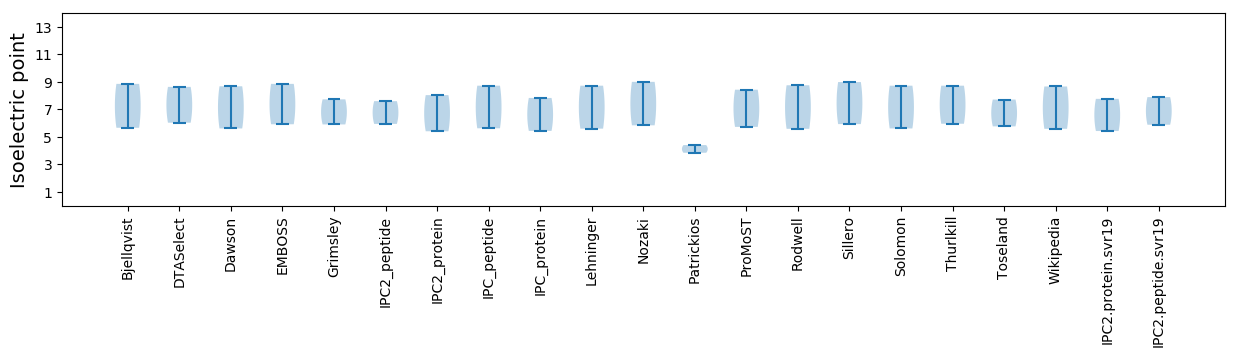
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
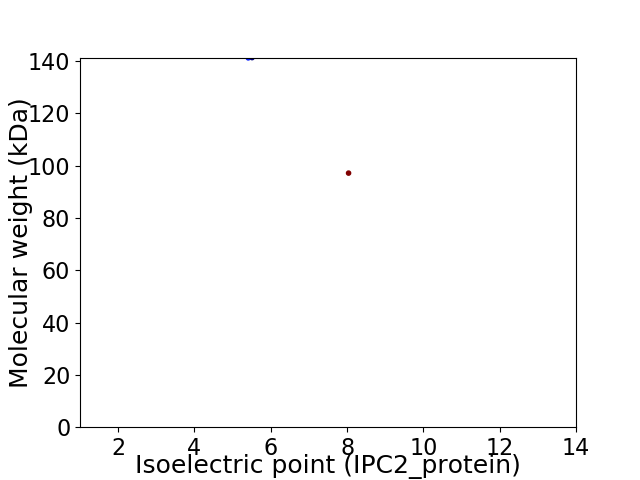
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
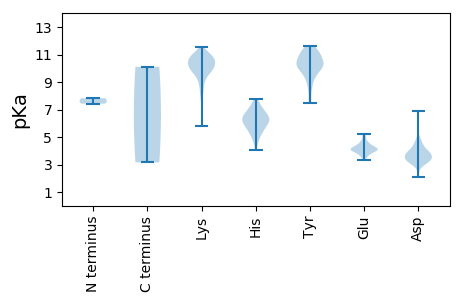
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KF81|A0A1L3KF81_9VIRU Uncharacterized protein OS=Beihai toti-like virus 4 OX=1922734 PE=4 SV=1
MM1 pKa = 7.43GNAGAKK7 pKa = 10.29GSGEE11 pKa = 4.31GVGSAIGGALTGVSPMLPFPLNLVTGLGGTAANLVGKK48 pKa = 9.91LVGDD52 pKa = 4.32DD53 pKa = 3.34PVVAQEE59 pKa = 4.4AFQQILQQNPQLANQVQVLPPGPQVPPALHH89 pKa = 6.23SQIQTTPSSVVTSGEE104 pKa = 4.0TTPQSNQSGGVEE116 pKa = 3.98GLLGHH121 pKa = 7.27LKK123 pKa = 9.88EE124 pKa = 4.29ANKK127 pKa = 10.14LQSFGPFTHH136 pKa = 7.42APTQLNQQIDD146 pKa = 3.72QLISHH151 pKa = 7.8LGQGHH156 pKa = 5.59LTKK159 pKa = 9.44PTMAQVPTTSQIAPNTSIANKK180 pKa = 10.34LGTTQLTADD189 pKa = 3.87VAEE192 pKa = 4.0QHH194 pKa = 5.26YY195 pKa = 9.99VKK197 pKa = 10.05PIEE200 pKa = 4.26SALVSGEE207 pKa = 3.81AGKK210 pKa = 10.88VGSTNPATNFALALQQGLFPQAQNTMKK237 pKa = 10.28TLEE240 pKa = 4.19NQNQEE245 pKa = 4.25TNIPRR250 pKa = 11.84PLMPGSQNQTEE261 pKa = 4.35LTNPKK266 pKa = 10.16VGTSTMLPLVPVNNARR282 pKa = 11.84QLNLLNGMNTVGGEE296 pKa = 4.03NNIVNSNLGTSTTSARR312 pKa = 11.84PTSFGGILKK321 pKa = 8.87DD322 pKa = 3.48TALDD326 pKa = 3.73VANVGAKK333 pKa = 9.85EE334 pKa = 3.94LGKK337 pKa = 8.49TAGNFLRR344 pKa = 11.84DD345 pKa = 4.0RR346 pKa = 11.84GPDD349 pKa = 3.29MLKK352 pKa = 10.28QGISFLGKK360 pKa = 8.17TLGGLFRR367 pKa = 11.84GKK369 pKa = 10.48LVGDD373 pKa = 4.35APLQPGTWSPGDD385 pKa = 3.88FNLLPNTNEE394 pKa = 3.76MFKK397 pKa = 10.49PGAIGTDD404 pKa = 3.13IVKK407 pKa = 9.87MIGDD411 pKa = 3.96APIDD415 pKa = 3.45GVMSNDD421 pKa = 3.21EE422 pKa = 3.64ASAAVKK428 pKa = 10.81AMTTSIAITLFAKK441 pKa = 10.35LVIIEE446 pKa = 4.23QTLEE450 pKa = 3.73TALPIIFSNNIRR462 pKa = 11.84QAAAQTARR470 pKa = 11.84QITVQNGMVRR480 pKa = 11.84LSQLLFNTDD489 pKa = 4.44LIGNYY494 pKa = 9.32YY495 pKa = 10.01FYY497 pKa = 11.17DD498 pKa = 5.09GIPVYY503 pKa = 9.6RR504 pKa = 11.84TNHH507 pKa = 6.31KK508 pKa = 9.41DD509 pKa = 2.8WHH511 pKa = 6.21VEE513 pKa = 4.01SPIYY517 pKa = 9.97TPTYY521 pKa = 8.55TGLVPISDD529 pKa = 4.32SPPLLGNVTYY539 pKa = 10.31EE540 pKa = 3.88ALKK543 pKa = 10.19PLLSGAPASEE553 pKa = 4.36YY554 pKa = 10.37PSTLRR559 pKa = 11.84IKK561 pKa = 9.04TRR563 pKa = 11.84RR564 pKa = 11.84TDD566 pKa = 2.81IDD568 pKa = 3.5ALLTISSQGTYY579 pKa = 10.36EE580 pKa = 3.99LTASAGYY587 pKa = 10.57SFVCPLVKK595 pKa = 10.64LLGYY599 pKa = 10.2QSAIGQLPGDD609 pKa = 4.18EE610 pKa = 4.23MTQYY614 pKa = 10.46MSGVYY619 pKa = 9.3PVINFSQNPQPGATIFPWRR638 pKa = 11.84SGSSDD643 pKa = 3.42LVNHH647 pKa = 5.93NTWKK651 pKa = 8.09WLKK654 pKa = 9.54VAWVTYY660 pKa = 8.98TEE662 pKa = 4.1FADD665 pKa = 4.86AIVGMRR671 pKa = 11.84APWLDD676 pKa = 3.13VFQPINWGITCAVVFISRR694 pKa = 11.84SEE696 pKa = 4.08MQDD699 pKa = 2.54GATNIVRR706 pKa = 11.84MLSHH710 pKa = 6.77IAYY713 pKa = 8.62PFVIFPRR720 pKa = 11.84EE721 pKa = 4.18VKK723 pKa = 9.07WVQAHH728 pKa = 6.3KK729 pKa = 10.97DD730 pKa = 3.19EE731 pKa = 4.81GFRR734 pKa = 11.84EE735 pKa = 4.16PVMNTTACWPHH746 pKa = 6.95IFSAQIEE753 pKa = 4.77GPDD756 pKa = 3.39QAILFVVTDD765 pKa = 4.27LSDD768 pKa = 3.45RR769 pKa = 11.84QAGFITLGIDD779 pKa = 3.25NMVEE783 pKa = 3.92VLTDD787 pKa = 4.0VITPNTPWQLDD798 pKa = 3.88EE799 pKa = 4.5EE800 pKa = 4.53QHH802 pKa = 4.85EE803 pKa = 4.55TRR805 pKa = 11.84HH806 pKa = 6.26PILMPIFSVINEE818 pKa = 4.06PARR821 pKa = 11.84FQAVIMSEE829 pKa = 3.36IRR831 pKa = 11.84RR832 pKa = 11.84WEE834 pKa = 3.98SYY836 pKa = 10.38YY837 pKa = 11.37GNNSDD842 pKa = 4.57RR843 pKa = 11.84SSAMRR848 pKa = 11.84WCAEE852 pKa = 3.42QSVRR856 pKa = 11.84FGQPKK861 pKa = 9.77NFCFTPTGTHH871 pKa = 6.74NIYY874 pKa = 9.89TGYY877 pKa = 9.18TYY879 pKa = 11.07NADD882 pKa = 3.67PSVSPYY888 pKa = 11.26GDD890 pKa = 3.4LSKK893 pKa = 11.18VFIMSEE899 pKa = 3.99DD900 pKa = 3.4GKK902 pKa = 10.61KK903 pKa = 10.08RR904 pKa = 11.84WMFANMLSTTPLMITPNILDD924 pKa = 4.05GYY926 pKa = 10.6FRR928 pKa = 11.84TPHH931 pKa = 6.05NEE933 pKa = 3.57YY934 pKa = 8.26TTFATTHH941 pKa = 6.27FLGQEE946 pKa = 3.98DD947 pKa = 4.33PVVNFLIEE955 pKa = 4.04RR956 pKa = 11.84GMVSPAEE963 pKa = 4.1EE964 pKa = 4.48YY965 pKa = 7.46PTVRR969 pKa = 11.84FTLAPKK975 pKa = 9.89NYY977 pKa = 10.26SAITEE982 pKa = 4.56LAQLMAFIMDD992 pKa = 3.77SSAQARR998 pKa = 11.84SVTVKK1003 pKa = 10.56DD1004 pKa = 3.79YY1005 pKa = 11.55YY1006 pKa = 11.06SYY1008 pKa = 11.18PNHH1011 pKa = 7.25RR1012 pKa = 11.84LNDD1015 pKa = 3.6DD1016 pKa = 3.51TNQNVVTRR1024 pKa = 11.84MALEE1028 pKa = 4.14KK1029 pKa = 10.75VFFPVGHH1036 pKa = 6.58AVCTNGIQKK1045 pKa = 10.38PSFSVNYY1052 pKa = 9.67QSLEE1056 pKa = 4.07YY1057 pKa = 10.86NHH1059 pKa = 6.75TFPLYY1064 pKa = 10.47FDD1066 pKa = 4.86AVWSFLGDD1074 pKa = 3.37GLTSAKK1080 pKa = 10.26NSTVGVFGRR1089 pKa = 11.84IPWALAEE1096 pKa = 4.07RR1097 pKa = 11.84WHH1099 pKa = 6.7PSFGIRR1105 pKa = 11.84DD1106 pKa = 3.95TNTNVYY1112 pKa = 10.22GLDD1115 pKa = 3.58VKK1117 pKa = 9.97ITEE1120 pKa = 4.24HH1121 pKa = 6.11MGVNGYY1127 pKa = 8.84EE1128 pKa = 3.94PHH1130 pKa = 6.64EE1131 pKa = 5.14LISMMSCDD1139 pKa = 3.81YY1140 pKa = 10.79DD1141 pKa = 3.84VQFIASLFRR1150 pKa = 11.84VSKK1153 pKa = 10.01FLSGLMVSSLQVNNPLWLYY1172 pKa = 10.94DD1173 pKa = 3.15SSTNYY1178 pKa = 9.97FYY1180 pKa = 11.59YY1181 pKa = 10.79LAGATLPLDD1190 pKa = 3.75SVVGRR1195 pKa = 11.84IIALSEE1201 pKa = 3.75AVGVINIPNFNLTPLPICGFVPTYY1225 pKa = 10.24NISGGGRR1232 pKa = 11.84KK1233 pKa = 8.81FRR1235 pKa = 11.84FAAIQPVDD1243 pKa = 3.71SFVNPSGSNDD1253 pKa = 3.31YY1254 pKa = 11.15QLFYY1258 pKa = 10.39PVKK1261 pKa = 8.31TQASLRR1267 pKa = 11.84TTIGYY1272 pKa = 9.55HH1273 pKa = 5.51EE1274 pKa = 5.19FGHH1277 pKa = 6.61PLFSDD1282 pKa = 4.54FTSNNPFAYY1291 pKa = 10.11
MM1 pKa = 7.43GNAGAKK7 pKa = 10.29GSGEE11 pKa = 4.31GVGSAIGGALTGVSPMLPFPLNLVTGLGGTAANLVGKK48 pKa = 9.91LVGDD52 pKa = 4.32DD53 pKa = 3.34PVVAQEE59 pKa = 4.4AFQQILQQNPQLANQVQVLPPGPQVPPALHH89 pKa = 6.23SQIQTTPSSVVTSGEE104 pKa = 4.0TTPQSNQSGGVEE116 pKa = 3.98GLLGHH121 pKa = 7.27LKK123 pKa = 9.88EE124 pKa = 4.29ANKK127 pKa = 10.14LQSFGPFTHH136 pKa = 7.42APTQLNQQIDD146 pKa = 3.72QLISHH151 pKa = 7.8LGQGHH156 pKa = 5.59LTKK159 pKa = 9.44PTMAQVPTTSQIAPNTSIANKK180 pKa = 10.34LGTTQLTADD189 pKa = 3.87VAEE192 pKa = 4.0QHH194 pKa = 5.26YY195 pKa = 9.99VKK197 pKa = 10.05PIEE200 pKa = 4.26SALVSGEE207 pKa = 3.81AGKK210 pKa = 10.88VGSTNPATNFALALQQGLFPQAQNTMKK237 pKa = 10.28TLEE240 pKa = 4.19NQNQEE245 pKa = 4.25TNIPRR250 pKa = 11.84PLMPGSQNQTEE261 pKa = 4.35LTNPKK266 pKa = 10.16VGTSTMLPLVPVNNARR282 pKa = 11.84QLNLLNGMNTVGGEE296 pKa = 4.03NNIVNSNLGTSTTSARR312 pKa = 11.84PTSFGGILKK321 pKa = 8.87DD322 pKa = 3.48TALDD326 pKa = 3.73VANVGAKK333 pKa = 9.85EE334 pKa = 3.94LGKK337 pKa = 8.49TAGNFLRR344 pKa = 11.84DD345 pKa = 4.0RR346 pKa = 11.84GPDD349 pKa = 3.29MLKK352 pKa = 10.28QGISFLGKK360 pKa = 8.17TLGGLFRR367 pKa = 11.84GKK369 pKa = 10.48LVGDD373 pKa = 4.35APLQPGTWSPGDD385 pKa = 3.88FNLLPNTNEE394 pKa = 3.76MFKK397 pKa = 10.49PGAIGTDD404 pKa = 3.13IVKK407 pKa = 9.87MIGDD411 pKa = 3.96APIDD415 pKa = 3.45GVMSNDD421 pKa = 3.21EE422 pKa = 3.64ASAAVKK428 pKa = 10.81AMTTSIAITLFAKK441 pKa = 10.35LVIIEE446 pKa = 4.23QTLEE450 pKa = 3.73TALPIIFSNNIRR462 pKa = 11.84QAAAQTARR470 pKa = 11.84QITVQNGMVRR480 pKa = 11.84LSQLLFNTDD489 pKa = 4.44LIGNYY494 pKa = 9.32YY495 pKa = 10.01FYY497 pKa = 11.17DD498 pKa = 5.09GIPVYY503 pKa = 9.6RR504 pKa = 11.84TNHH507 pKa = 6.31KK508 pKa = 9.41DD509 pKa = 2.8WHH511 pKa = 6.21VEE513 pKa = 4.01SPIYY517 pKa = 9.97TPTYY521 pKa = 8.55TGLVPISDD529 pKa = 4.32SPPLLGNVTYY539 pKa = 10.31EE540 pKa = 3.88ALKK543 pKa = 10.19PLLSGAPASEE553 pKa = 4.36YY554 pKa = 10.37PSTLRR559 pKa = 11.84IKK561 pKa = 9.04TRR563 pKa = 11.84RR564 pKa = 11.84TDD566 pKa = 2.81IDD568 pKa = 3.5ALLTISSQGTYY579 pKa = 10.36EE580 pKa = 3.99LTASAGYY587 pKa = 10.57SFVCPLVKK595 pKa = 10.64LLGYY599 pKa = 10.2QSAIGQLPGDD609 pKa = 4.18EE610 pKa = 4.23MTQYY614 pKa = 10.46MSGVYY619 pKa = 9.3PVINFSQNPQPGATIFPWRR638 pKa = 11.84SGSSDD643 pKa = 3.42LVNHH647 pKa = 5.93NTWKK651 pKa = 8.09WLKK654 pKa = 9.54VAWVTYY660 pKa = 8.98TEE662 pKa = 4.1FADD665 pKa = 4.86AIVGMRR671 pKa = 11.84APWLDD676 pKa = 3.13VFQPINWGITCAVVFISRR694 pKa = 11.84SEE696 pKa = 4.08MQDD699 pKa = 2.54GATNIVRR706 pKa = 11.84MLSHH710 pKa = 6.77IAYY713 pKa = 8.62PFVIFPRR720 pKa = 11.84EE721 pKa = 4.18VKK723 pKa = 9.07WVQAHH728 pKa = 6.3KK729 pKa = 10.97DD730 pKa = 3.19EE731 pKa = 4.81GFRR734 pKa = 11.84EE735 pKa = 4.16PVMNTTACWPHH746 pKa = 6.95IFSAQIEE753 pKa = 4.77GPDD756 pKa = 3.39QAILFVVTDD765 pKa = 4.27LSDD768 pKa = 3.45RR769 pKa = 11.84QAGFITLGIDD779 pKa = 3.25NMVEE783 pKa = 3.92VLTDD787 pKa = 4.0VITPNTPWQLDD798 pKa = 3.88EE799 pKa = 4.5EE800 pKa = 4.53QHH802 pKa = 4.85EE803 pKa = 4.55TRR805 pKa = 11.84HH806 pKa = 6.26PILMPIFSVINEE818 pKa = 4.06PARR821 pKa = 11.84FQAVIMSEE829 pKa = 3.36IRR831 pKa = 11.84RR832 pKa = 11.84WEE834 pKa = 3.98SYY836 pKa = 10.38YY837 pKa = 11.37GNNSDD842 pKa = 4.57RR843 pKa = 11.84SSAMRR848 pKa = 11.84WCAEE852 pKa = 3.42QSVRR856 pKa = 11.84FGQPKK861 pKa = 9.77NFCFTPTGTHH871 pKa = 6.74NIYY874 pKa = 9.89TGYY877 pKa = 9.18TYY879 pKa = 11.07NADD882 pKa = 3.67PSVSPYY888 pKa = 11.26GDD890 pKa = 3.4LSKK893 pKa = 11.18VFIMSEE899 pKa = 3.99DD900 pKa = 3.4GKK902 pKa = 10.61KK903 pKa = 10.08RR904 pKa = 11.84WMFANMLSTTPLMITPNILDD924 pKa = 4.05GYY926 pKa = 10.6FRR928 pKa = 11.84TPHH931 pKa = 6.05NEE933 pKa = 3.57YY934 pKa = 8.26TTFATTHH941 pKa = 6.27FLGQEE946 pKa = 3.98DD947 pKa = 4.33PVVNFLIEE955 pKa = 4.04RR956 pKa = 11.84GMVSPAEE963 pKa = 4.1EE964 pKa = 4.48YY965 pKa = 7.46PTVRR969 pKa = 11.84FTLAPKK975 pKa = 9.89NYY977 pKa = 10.26SAITEE982 pKa = 4.56LAQLMAFIMDD992 pKa = 3.77SSAQARR998 pKa = 11.84SVTVKK1003 pKa = 10.56DD1004 pKa = 3.79YY1005 pKa = 11.55YY1006 pKa = 11.06SYY1008 pKa = 11.18PNHH1011 pKa = 7.25RR1012 pKa = 11.84LNDD1015 pKa = 3.6DD1016 pKa = 3.51TNQNVVTRR1024 pKa = 11.84MALEE1028 pKa = 4.14KK1029 pKa = 10.75VFFPVGHH1036 pKa = 6.58AVCTNGIQKK1045 pKa = 10.38PSFSVNYY1052 pKa = 9.67QSLEE1056 pKa = 4.07YY1057 pKa = 10.86NHH1059 pKa = 6.75TFPLYY1064 pKa = 10.47FDD1066 pKa = 4.86AVWSFLGDD1074 pKa = 3.37GLTSAKK1080 pKa = 10.26NSTVGVFGRR1089 pKa = 11.84IPWALAEE1096 pKa = 4.07RR1097 pKa = 11.84WHH1099 pKa = 6.7PSFGIRR1105 pKa = 11.84DD1106 pKa = 3.95TNTNVYY1112 pKa = 10.22GLDD1115 pKa = 3.58VKK1117 pKa = 9.97ITEE1120 pKa = 4.24HH1121 pKa = 6.11MGVNGYY1127 pKa = 8.84EE1128 pKa = 3.94PHH1130 pKa = 6.64EE1131 pKa = 5.14LISMMSCDD1139 pKa = 3.81YY1140 pKa = 10.79DD1141 pKa = 3.84VQFIASLFRR1150 pKa = 11.84VSKK1153 pKa = 10.01FLSGLMVSSLQVNNPLWLYY1172 pKa = 10.94DD1173 pKa = 3.15SSTNYY1178 pKa = 9.97FYY1180 pKa = 11.59YY1181 pKa = 10.79LAGATLPLDD1190 pKa = 3.75SVVGRR1195 pKa = 11.84IIALSEE1201 pKa = 3.75AVGVINIPNFNLTPLPICGFVPTYY1225 pKa = 10.24NISGGGRR1232 pKa = 11.84KK1233 pKa = 8.81FRR1235 pKa = 11.84FAAIQPVDD1243 pKa = 3.71SFVNPSGSNDD1253 pKa = 3.31YY1254 pKa = 11.15QLFYY1258 pKa = 10.39PVKK1261 pKa = 8.31TQASLRR1267 pKa = 11.84TTIGYY1272 pKa = 9.55HH1273 pKa = 5.51EE1274 pKa = 5.19FGHH1277 pKa = 6.61PLFSDD1282 pKa = 4.54FTSNNPFAYY1291 pKa = 10.11
Molecular weight: 141.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF81|A0A1L3KF81_9VIRU Uncharacterized protein OS=Beihai toti-like virus 4 OX=1922734 PE=4 SV=1
MM1 pKa = 7.82AEE3 pKa = 3.9NLKK6 pKa = 10.27DD7 pKa = 3.68IKK9 pKa = 10.91VLYY12 pKa = 9.19PDD14 pKa = 3.95YY15 pKa = 11.49EE16 pKa = 4.43MTEE19 pKa = 4.3SMWLDD24 pKa = 3.11IVNIKK29 pKa = 10.14LDD31 pKa = 3.94KK32 pKa = 11.26NDD34 pKa = 3.8FLNDD38 pKa = 3.35LTEE41 pKa = 4.62DD42 pKa = 3.83SNVPGISYY50 pKa = 9.93FSSYY54 pKa = 9.84HH55 pKa = 4.79VNRR58 pKa = 11.84IRR60 pKa = 11.84QMPPMYY66 pKa = 10.44FEE68 pKa = 5.16QEE70 pKa = 4.1FKK72 pKa = 11.01NYY74 pKa = 9.92PLTLKK79 pKa = 10.69FCFSLLLQLHH89 pKa = 6.07PVYY92 pKa = 10.41RR93 pKa = 11.84DD94 pKa = 3.3IIISVLYY101 pKa = 10.26AVNILPNCLTVTKK114 pKa = 10.49YY115 pKa = 10.63IEE117 pKa = 4.16VCKK120 pKa = 10.77KK121 pKa = 8.24LTSWAKK127 pKa = 10.0KK128 pKa = 9.74CNSLASRR135 pKa = 11.84LINNKK140 pKa = 9.09FNCNVEE146 pKa = 4.3DD147 pKa = 4.35KK148 pKa = 10.84FWMILVDD155 pKa = 3.37WDD157 pKa = 3.54IFSGYY162 pKa = 7.79NFKK165 pKa = 11.22YY166 pKa = 10.43NEE168 pKa = 4.13SKK170 pKa = 10.64IIDD173 pKa = 4.21SIKK176 pKa = 9.86EE177 pKa = 3.83WGSGDD182 pKa = 3.08IYY184 pKa = 10.91KK185 pKa = 10.96GNNEE189 pKa = 3.98VSSDD193 pKa = 3.32KK194 pKa = 10.9YY195 pKa = 9.7YY196 pKa = 11.52KK197 pKa = 10.89LMEE200 pKa = 4.66KK201 pKa = 10.58YY202 pKa = 10.15VDD204 pKa = 3.42KK205 pKa = 11.01LLEE208 pKa = 4.48FGTLEE213 pKa = 4.09KK214 pKa = 11.01KK215 pKa = 10.69NILSLRR221 pKa = 11.84DD222 pKa = 3.55WLSDD226 pKa = 3.08PTKK229 pKa = 9.86WVTAGSSNGVRR240 pKa = 11.84VDD242 pKa = 2.96IFSYY246 pKa = 10.84DD247 pKa = 2.92KK248 pKa = 11.11NRR250 pKa = 11.84NVKK253 pKa = 9.69SAATKK258 pKa = 10.07SVFSVKK264 pKa = 10.44YY265 pKa = 10.63DD266 pKa = 3.09VDD268 pKa = 3.88DD269 pKa = 4.4LYY271 pKa = 11.38KK272 pKa = 10.47ILMTNTKK279 pKa = 9.43PQDD282 pKa = 3.2SRR284 pKa = 11.84VNIKK288 pKa = 10.75LEE290 pKa = 4.22AGQKK294 pKa = 9.74NRR296 pKa = 11.84IIVSSGWVNQLRR308 pKa = 11.84MAYY311 pKa = 9.3ISSWLEE317 pKa = 3.51KK318 pKa = 10.44SLKK321 pKa = 9.16NVPWTTLFSTNQDD334 pKa = 2.1AWTRR338 pKa = 11.84NVRR341 pKa = 11.84KK342 pKa = 9.67CSHH345 pKa = 6.47IKK347 pKa = 10.15SGSIMFPLDD356 pKa = 3.87AKK358 pKa = 11.16NFDD361 pKa = 3.73QNVSKK366 pKa = 11.57GEE368 pKa = 3.67IDD370 pKa = 3.47VCFNSIRR377 pKa = 11.84KK378 pKa = 9.15IINKK382 pKa = 9.54SLAGEE387 pKa = 4.11KK388 pKa = 10.5RR389 pKa = 11.84FDD391 pKa = 3.74CLKK394 pKa = 10.9LIDD397 pKa = 4.83IIQNQFWNTFVVYY410 pKa = 10.87KK411 pKa = 9.67NMRR414 pKa = 11.84MLWEE418 pKa = 4.66HH419 pKa = 6.24GMPSGIRR426 pKa = 11.84WTAFLDD432 pKa = 3.66TLINAARR439 pKa = 11.84VCIINDD445 pKa = 3.37ILSKK449 pKa = 11.0GSTIPIKK456 pKa = 10.42EE457 pKa = 3.91YY458 pKa = 9.7TAQGDD463 pKa = 3.86DD464 pKa = 4.69DD465 pKa = 4.7DD466 pKa = 6.89LEE468 pKa = 4.23VSKK471 pKa = 10.67WDD473 pKa = 4.66DD474 pKa = 3.02ILNIFNTYY482 pKa = 9.82NSLGIVLAPQKK493 pKa = 10.87NFVSTKK499 pKa = 5.83MTEE502 pKa = 4.24FLRR505 pKa = 11.84KK506 pKa = 9.83VYY508 pKa = 10.44FSDD511 pKa = 5.82LIMGYY516 pKa = 7.53ITRR519 pKa = 11.84KK520 pKa = 7.68MVNVCFVNPEE530 pKa = 4.15KK531 pKa = 11.19NLVNYY536 pKa = 10.28RR537 pKa = 11.84DD538 pKa = 3.65VDD540 pKa = 3.6LHH542 pKa = 5.94TIRR545 pKa = 11.84DD546 pKa = 3.15IWLEE550 pKa = 3.71LMQRR554 pKa = 11.84GGDD557 pKa = 3.72FEE559 pKa = 4.64YY560 pKa = 11.16CEE562 pKa = 4.44SKK564 pKa = 10.8LIEE567 pKa = 4.98HH568 pKa = 7.09IALVLFNKK576 pKa = 10.07LGKK579 pKa = 9.86IEE581 pKa = 4.94GYY583 pKa = 10.47SKK585 pKa = 10.37LTDD588 pKa = 3.08RR589 pKa = 11.84HH590 pKa = 5.42LAIAKK595 pKa = 9.81DD596 pKa = 3.95VLSTPASLGGLGCLWFGTRR615 pKa = 11.84TWRR618 pKa = 11.84VLQEE622 pKa = 3.99KK623 pKa = 10.13RR624 pKa = 11.84SEE626 pKa = 3.53RR627 pKa = 11.84HH628 pKa = 4.05YY629 pKa = 10.64QLRR632 pKa = 11.84GPLGVWTKK640 pKa = 10.81VLSATNSVTGNVRR653 pKa = 11.84RR654 pKa = 11.84EE655 pKa = 3.77NEE657 pKa = 3.54AVIRR661 pKa = 11.84DD662 pKa = 4.25KK663 pKa = 10.82IVQTLIPKK671 pKa = 9.67HH672 pKa = 5.54EE673 pKa = 4.14RR674 pKa = 11.84VPRR677 pKa = 11.84IIEE680 pKa = 4.27CEE682 pKa = 3.77LLEE685 pKa = 4.52IPFSGLIQVPEE696 pKa = 4.17ITPSLQRR703 pKa = 11.84TLPVWRR709 pKa = 11.84MRR711 pKa = 11.84DD712 pKa = 3.49DD713 pKa = 3.54IGPVDD718 pKa = 4.19VNCVLNACKK727 pKa = 10.25EE728 pKa = 4.02NRR730 pKa = 11.84DD731 pKa = 3.34WGLLRR736 pKa = 11.84SITSEE741 pKa = 4.18SVLGTFDD748 pKa = 4.02NMLSSKK754 pKa = 10.27HH755 pKa = 5.71KK756 pKa = 10.56RR757 pKa = 11.84EE758 pKa = 4.34LLITWVMDD766 pKa = 3.24DD767 pKa = 3.36WTVNAPSHH775 pKa = 5.98RR776 pKa = 11.84VWGKK780 pKa = 7.23TALSAMFVGWKK791 pKa = 9.7NYY793 pKa = 9.94CINRR797 pKa = 11.84WYY799 pKa = 10.12EE800 pKa = 3.88VNNKK804 pKa = 8.83GRR806 pKa = 11.84DD807 pKa = 3.9YY808 pKa = 9.93YY809 pKa = 11.17TKK811 pKa = 10.37LCVSSEE817 pKa = 4.22YY818 pKa = 11.18NFYY821 pKa = 11.02KK822 pKa = 10.58YY823 pKa = 11.18VNTHH827 pKa = 5.02EE828 pKa = 4.95HH829 pKa = 5.53PVVMRR834 pKa = 11.84FF835 pKa = 3.16
MM1 pKa = 7.82AEE3 pKa = 3.9NLKK6 pKa = 10.27DD7 pKa = 3.68IKK9 pKa = 10.91VLYY12 pKa = 9.19PDD14 pKa = 3.95YY15 pKa = 11.49EE16 pKa = 4.43MTEE19 pKa = 4.3SMWLDD24 pKa = 3.11IVNIKK29 pKa = 10.14LDD31 pKa = 3.94KK32 pKa = 11.26NDD34 pKa = 3.8FLNDD38 pKa = 3.35LTEE41 pKa = 4.62DD42 pKa = 3.83SNVPGISYY50 pKa = 9.93FSSYY54 pKa = 9.84HH55 pKa = 4.79VNRR58 pKa = 11.84IRR60 pKa = 11.84QMPPMYY66 pKa = 10.44FEE68 pKa = 5.16QEE70 pKa = 4.1FKK72 pKa = 11.01NYY74 pKa = 9.92PLTLKK79 pKa = 10.69FCFSLLLQLHH89 pKa = 6.07PVYY92 pKa = 10.41RR93 pKa = 11.84DD94 pKa = 3.3IIISVLYY101 pKa = 10.26AVNILPNCLTVTKK114 pKa = 10.49YY115 pKa = 10.63IEE117 pKa = 4.16VCKK120 pKa = 10.77KK121 pKa = 8.24LTSWAKK127 pKa = 10.0KK128 pKa = 9.74CNSLASRR135 pKa = 11.84LINNKK140 pKa = 9.09FNCNVEE146 pKa = 4.3DD147 pKa = 4.35KK148 pKa = 10.84FWMILVDD155 pKa = 3.37WDD157 pKa = 3.54IFSGYY162 pKa = 7.79NFKK165 pKa = 11.22YY166 pKa = 10.43NEE168 pKa = 4.13SKK170 pKa = 10.64IIDD173 pKa = 4.21SIKK176 pKa = 9.86EE177 pKa = 3.83WGSGDD182 pKa = 3.08IYY184 pKa = 10.91KK185 pKa = 10.96GNNEE189 pKa = 3.98VSSDD193 pKa = 3.32KK194 pKa = 10.9YY195 pKa = 9.7YY196 pKa = 11.52KK197 pKa = 10.89LMEE200 pKa = 4.66KK201 pKa = 10.58YY202 pKa = 10.15VDD204 pKa = 3.42KK205 pKa = 11.01LLEE208 pKa = 4.48FGTLEE213 pKa = 4.09KK214 pKa = 11.01KK215 pKa = 10.69NILSLRR221 pKa = 11.84DD222 pKa = 3.55WLSDD226 pKa = 3.08PTKK229 pKa = 9.86WVTAGSSNGVRR240 pKa = 11.84VDD242 pKa = 2.96IFSYY246 pKa = 10.84DD247 pKa = 2.92KK248 pKa = 11.11NRR250 pKa = 11.84NVKK253 pKa = 9.69SAATKK258 pKa = 10.07SVFSVKK264 pKa = 10.44YY265 pKa = 10.63DD266 pKa = 3.09VDD268 pKa = 3.88DD269 pKa = 4.4LYY271 pKa = 11.38KK272 pKa = 10.47ILMTNTKK279 pKa = 9.43PQDD282 pKa = 3.2SRR284 pKa = 11.84VNIKK288 pKa = 10.75LEE290 pKa = 4.22AGQKK294 pKa = 9.74NRR296 pKa = 11.84IIVSSGWVNQLRR308 pKa = 11.84MAYY311 pKa = 9.3ISSWLEE317 pKa = 3.51KK318 pKa = 10.44SLKK321 pKa = 9.16NVPWTTLFSTNQDD334 pKa = 2.1AWTRR338 pKa = 11.84NVRR341 pKa = 11.84KK342 pKa = 9.67CSHH345 pKa = 6.47IKK347 pKa = 10.15SGSIMFPLDD356 pKa = 3.87AKK358 pKa = 11.16NFDD361 pKa = 3.73QNVSKK366 pKa = 11.57GEE368 pKa = 3.67IDD370 pKa = 3.47VCFNSIRR377 pKa = 11.84KK378 pKa = 9.15IINKK382 pKa = 9.54SLAGEE387 pKa = 4.11KK388 pKa = 10.5RR389 pKa = 11.84FDD391 pKa = 3.74CLKK394 pKa = 10.9LIDD397 pKa = 4.83IIQNQFWNTFVVYY410 pKa = 10.87KK411 pKa = 9.67NMRR414 pKa = 11.84MLWEE418 pKa = 4.66HH419 pKa = 6.24GMPSGIRR426 pKa = 11.84WTAFLDD432 pKa = 3.66TLINAARR439 pKa = 11.84VCIINDD445 pKa = 3.37ILSKK449 pKa = 11.0GSTIPIKK456 pKa = 10.42EE457 pKa = 3.91YY458 pKa = 9.7TAQGDD463 pKa = 3.86DD464 pKa = 4.69DD465 pKa = 4.7DD466 pKa = 6.89LEE468 pKa = 4.23VSKK471 pKa = 10.67WDD473 pKa = 4.66DD474 pKa = 3.02ILNIFNTYY482 pKa = 9.82NSLGIVLAPQKK493 pKa = 10.87NFVSTKK499 pKa = 5.83MTEE502 pKa = 4.24FLRR505 pKa = 11.84KK506 pKa = 9.83VYY508 pKa = 10.44FSDD511 pKa = 5.82LIMGYY516 pKa = 7.53ITRR519 pKa = 11.84KK520 pKa = 7.68MVNVCFVNPEE530 pKa = 4.15KK531 pKa = 11.19NLVNYY536 pKa = 10.28RR537 pKa = 11.84DD538 pKa = 3.65VDD540 pKa = 3.6LHH542 pKa = 5.94TIRR545 pKa = 11.84DD546 pKa = 3.15IWLEE550 pKa = 3.71LMQRR554 pKa = 11.84GGDD557 pKa = 3.72FEE559 pKa = 4.64YY560 pKa = 11.16CEE562 pKa = 4.44SKK564 pKa = 10.8LIEE567 pKa = 4.98HH568 pKa = 7.09IALVLFNKK576 pKa = 10.07LGKK579 pKa = 9.86IEE581 pKa = 4.94GYY583 pKa = 10.47SKK585 pKa = 10.37LTDD588 pKa = 3.08RR589 pKa = 11.84HH590 pKa = 5.42LAIAKK595 pKa = 9.81DD596 pKa = 3.95VLSTPASLGGLGCLWFGTRR615 pKa = 11.84TWRR618 pKa = 11.84VLQEE622 pKa = 3.99KK623 pKa = 10.13RR624 pKa = 11.84SEE626 pKa = 3.53RR627 pKa = 11.84HH628 pKa = 4.05YY629 pKa = 10.64QLRR632 pKa = 11.84GPLGVWTKK640 pKa = 10.81VLSATNSVTGNVRR653 pKa = 11.84RR654 pKa = 11.84EE655 pKa = 3.77NEE657 pKa = 3.54AVIRR661 pKa = 11.84DD662 pKa = 4.25KK663 pKa = 10.82IVQTLIPKK671 pKa = 9.67HH672 pKa = 5.54EE673 pKa = 4.14RR674 pKa = 11.84VPRR677 pKa = 11.84IIEE680 pKa = 4.27CEE682 pKa = 3.77LLEE685 pKa = 4.52IPFSGLIQVPEE696 pKa = 4.17ITPSLQRR703 pKa = 11.84TLPVWRR709 pKa = 11.84MRR711 pKa = 11.84DD712 pKa = 3.49DD713 pKa = 3.54IGPVDD718 pKa = 4.19VNCVLNACKK727 pKa = 10.25EE728 pKa = 4.02NRR730 pKa = 11.84DD731 pKa = 3.34WGLLRR736 pKa = 11.84SITSEE741 pKa = 4.18SVLGTFDD748 pKa = 4.02NMLSSKK754 pKa = 10.27HH755 pKa = 5.71KK756 pKa = 10.56RR757 pKa = 11.84EE758 pKa = 4.34LLITWVMDD766 pKa = 3.24DD767 pKa = 3.36WTVNAPSHH775 pKa = 5.98RR776 pKa = 11.84VWGKK780 pKa = 7.23TALSAMFVGWKK791 pKa = 9.7NYY793 pKa = 9.94CINRR797 pKa = 11.84WYY799 pKa = 10.12EE800 pKa = 3.88VNNKK804 pKa = 8.83GRR806 pKa = 11.84DD807 pKa = 3.9YY808 pKa = 9.93YY809 pKa = 11.17TKK811 pKa = 10.37LCVSSEE817 pKa = 4.22YY818 pKa = 11.18NFYY821 pKa = 11.02KK822 pKa = 10.58YY823 pKa = 11.18VNTHH827 pKa = 5.02EE828 pKa = 4.95HH829 pKa = 5.53PVVMRR834 pKa = 11.84FF835 pKa = 3.16
Molecular weight: 97.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2126 |
835 |
1291 |
1063.0 |
119.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.597 ± 1.408 | 1.176 ± 0.512 |
4.986 ± 0.811 | 4.421 ± 0.434 |
4.563 ± 0.292 | 6.585 ± 1.283 |
1.787 ± 0.137 | 6.397 ± 0.684 |
5.127 ± 1.797 | 8.984 ± 0.356 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.681 ± 0.028 | 6.726 ± 0.345 |
5.503 ± 1.424 | 4.045 ± 1.126 |
3.998 ± 0.686 | 7.291 ± 0.08 |
7.008 ± 1.107 | 7.338 ± 0.195 |
2.07 ± 0.622 | 3.716 ± 0.283 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
