
Phenylobacterium kunshanense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Phenylobacterium
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
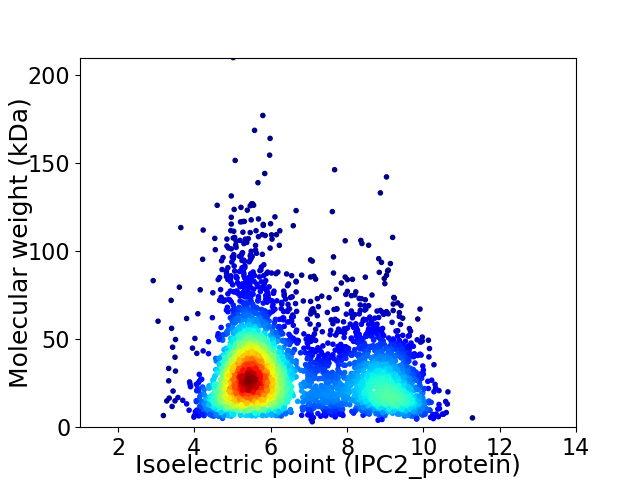
Virtual 2D-PAGE plot for 4047 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A328BJ08|A0A328BJ08_9CAUL LprI domain-containing protein OS=Phenylobacterium kunshanense OX=1445034 GN=DJ019_05995 PE=4 SV=1
MM1 pKa = 7.69PFASATDD8 pKa = 3.65TAQAATDD15 pKa = 3.64IFAQGVRR22 pKa = 11.84PFGGEE27 pKa = 3.72APLGCACPVCRR38 pKa = 11.84GAVTVVEE45 pKa = 4.77DD46 pKa = 3.88GGEE49 pKa = 4.17GALQGYY55 pKa = 9.74LNADD59 pKa = 3.35QRR61 pKa = 11.84SGAVVNGKK69 pKa = 8.97QSFSIDD75 pKa = 3.22RR76 pKa = 11.84AGLQMTGFDD85 pKa = 4.87PDD87 pKa = 3.33TMQPYY92 pKa = 9.39PGWGGTAGQAFTVTYY107 pKa = 9.95AFRR110 pKa = 11.84SSAPFNMPSDD120 pKa = 3.68TEE122 pKa = 4.06GFQRR126 pKa = 11.84FNAQQIYY133 pKa = 9.09QAEE136 pKa = 4.15QALAAWSDD144 pKa = 3.54VANITFVRR152 pKa = 11.84VGSGAEE158 pKa = 3.99GEE160 pKa = 4.15EE161 pKa = 4.37AYY163 pKa = 11.16SNNAAILFSNYY174 pKa = 8.42TSGEE178 pKa = 4.25SGSSAFASYY187 pKa = 10.04PGSTAASSTAGDD199 pKa = 3.19VWINITAGSNSIPSMGNYY217 pKa = 7.96GAHH220 pKa = 5.43VLIHH224 pKa = 6.69EE225 pKa = 4.71IGHH228 pKa = 6.91AIGLAHH234 pKa = 7.03PGDD237 pKa = 4.39YY238 pKa = 11.02NSTADD243 pKa = 4.0GGAITYY249 pKa = 10.65SSDD252 pKa = 2.82AEE254 pKa = 4.36YY255 pKa = 11.5YY256 pKa = 10.46EE257 pKa = 4.41DD258 pKa = 3.55TRR260 pKa = 11.84QYY262 pKa = 10.95TVMSYY267 pKa = 10.54FSEE270 pKa = 4.7SNTGASYY277 pKa = 10.61GGRR280 pKa = 11.84YY281 pKa = 9.44ASAPQLDD288 pKa = 5.29DD289 pKa = 3.1IRR291 pKa = 11.84AAQIEE296 pKa = 4.67YY297 pKa = 8.86GANMTTRR304 pKa = 11.84TGDD307 pKa = 3.13TVYY310 pKa = 10.98GFNANAGRR318 pKa = 11.84DD319 pKa = 3.38WFAATSSSTRR329 pKa = 11.84LIFAVWDD336 pKa = 3.62AGGNDD341 pKa = 3.32TFDD344 pKa = 3.72FSGYY348 pKa = 8.77SQNQTIDD355 pKa = 3.2LRR357 pKa = 11.84EE358 pKa = 4.04GFFSNVGSLVGNVAVAQGAKK378 pKa = 9.6IEE380 pKa = 4.01NAIGGFGADD389 pKa = 4.01TINGNALANRR399 pKa = 11.84IAGNAGADD407 pKa = 3.93RR408 pKa = 11.84IFSGAGNDD416 pKa = 4.08TIEE419 pKa = 4.48GGAGSGYY426 pKa = 10.57LRR428 pKa = 11.84GEE430 pKa = 4.79DD431 pKa = 4.1GNDD434 pKa = 3.73SIIGGAAFDD443 pKa = 5.6DD444 pKa = 3.58IHH446 pKa = 7.7GNTGNDD452 pKa = 3.43TASGGDD458 pKa = 3.33GDD460 pKa = 4.18DD461 pKa = 3.26WVVGGKK467 pKa = 10.34DD468 pKa = 3.3NDD470 pKa = 3.87VLFGDD475 pKa = 3.87AGGDD479 pKa = 3.51VVFGNLGDD487 pKa = 3.97DD488 pKa = 3.99TVHH491 pKa = 6.6GGSGNDD497 pKa = 3.53TVRR500 pKa = 11.84GGQGNDD506 pKa = 3.22TLSGGDD512 pKa = 3.25GDD514 pKa = 5.68DD515 pKa = 3.66YY516 pKa = 11.8VSGDD520 pKa = 3.55RR521 pKa = 11.84GSDD524 pKa = 3.11TMTGGAGADD533 pKa = 3.72LFHH536 pKa = 6.89TSSDD540 pKa = 3.28AGIDD544 pKa = 3.61SVFDD548 pKa = 3.87FNLAEE553 pKa = 4.47GDD555 pKa = 4.04RR556 pKa = 11.84VLLDD560 pKa = 3.81PGTTFSVAQVGADD573 pKa = 3.45VVITLGSPSDD583 pKa = 3.56QMILVGIQLSSLTPGWIFGAA603 pKa = 4.2
MM1 pKa = 7.69PFASATDD8 pKa = 3.65TAQAATDD15 pKa = 3.64IFAQGVRR22 pKa = 11.84PFGGEE27 pKa = 3.72APLGCACPVCRR38 pKa = 11.84GAVTVVEE45 pKa = 4.77DD46 pKa = 3.88GGEE49 pKa = 4.17GALQGYY55 pKa = 9.74LNADD59 pKa = 3.35QRR61 pKa = 11.84SGAVVNGKK69 pKa = 8.97QSFSIDD75 pKa = 3.22RR76 pKa = 11.84AGLQMTGFDD85 pKa = 4.87PDD87 pKa = 3.33TMQPYY92 pKa = 9.39PGWGGTAGQAFTVTYY107 pKa = 9.95AFRR110 pKa = 11.84SSAPFNMPSDD120 pKa = 3.68TEE122 pKa = 4.06GFQRR126 pKa = 11.84FNAQQIYY133 pKa = 9.09QAEE136 pKa = 4.15QALAAWSDD144 pKa = 3.54VANITFVRR152 pKa = 11.84VGSGAEE158 pKa = 3.99GEE160 pKa = 4.15EE161 pKa = 4.37AYY163 pKa = 11.16SNNAAILFSNYY174 pKa = 8.42TSGEE178 pKa = 4.25SGSSAFASYY187 pKa = 10.04PGSTAASSTAGDD199 pKa = 3.19VWINITAGSNSIPSMGNYY217 pKa = 7.96GAHH220 pKa = 5.43VLIHH224 pKa = 6.69EE225 pKa = 4.71IGHH228 pKa = 6.91AIGLAHH234 pKa = 7.03PGDD237 pKa = 4.39YY238 pKa = 11.02NSTADD243 pKa = 4.0GGAITYY249 pKa = 10.65SSDD252 pKa = 2.82AEE254 pKa = 4.36YY255 pKa = 11.5YY256 pKa = 10.46EE257 pKa = 4.41DD258 pKa = 3.55TRR260 pKa = 11.84QYY262 pKa = 10.95TVMSYY267 pKa = 10.54FSEE270 pKa = 4.7SNTGASYY277 pKa = 10.61GGRR280 pKa = 11.84YY281 pKa = 9.44ASAPQLDD288 pKa = 5.29DD289 pKa = 3.1IRR291 pKa = 11.84AAQIEE296 pKa = 4.67YY297 pKa = 8.86GANMTTRR304 pKa = 11.84TGDD307 pKa = 3.13TVYY310 pKa = 10.98GFNANAGRR318 pKa = 11.84DD319 pKa = 3.38WFAATSSSTRR329 pKa = 11.84LIFAVWDD336 pKa = 3.62AGGNDD341 pKa = 3.32TFDD344 pKa = 3.72FSGYY348 pKa = 8.77SQNQTIDD355 pKa = 3.2LRR357 pKa = 11.84EE358 pKa = 4.04GFFSNVGSLVGNVAVAQGAKK378 pKa = 9.6IEE380 pKa = 4.01NAIGGFGADD389 pKa = 4.01TINGNALANRR399 pKa = 11.84IAGNAGADD407 pKa = 3.93RR408 pKa = 11.84IFSGAGNDD416 pKa = 4.08TIEE419 pKa = 4.48GGAGSGYY426 pKa = 10.57LRR428 pKa = 11.84GEE430 pKa = 4.79DD431 pKa = 4.1GNDD434 pKa = 3.73SIIGGAAFDD443 pKa = 5.6DD444 pKa = 3.58IHH446 pKa = 7.7GNTGNDD452 pKa = 3.43TASGGDD458 pKa = 3.33GDD460 pKa = 4.18DD461 pKa = 3.26WVVGGKK467 pKa = 10.34DD468 pKa = 3.3NDD470 pKa = 3.87VLFGDD475 pKa = 3.87AGGDD479 pKa = 3.51VVFGNLGDD487 pKa = 3.97DD488 pKa = 3.99TVHH491 pKa = 6.6GGSGNDD497 pKa = 3.53TVRR500 pKa = 11.84GGQGNDD506 pKa = 3.22TLSGGDD512 pKa = 3.25GDD514 pKa = 5.68DD515 pKa = 3.66YY516 pKa = 11.8VSGDD520 pKa = 3.55RR521 pKa = 11.84GSDD524 pKa = 3.11TMTGGAGADD533 pKa = 3.72LFHH536 pKa = 6.89TSSDD540 pKa = 3.28AGIDD544 pKa = 3.61SVFDD548 pKa = 3.87FNLAEE553 pKa = 4.47GDD555 pKa = 4.04RR556 pKa = 11.84VLLDD560 pKa = 3.81PGTTFSVAQVGADD573 pKa = 3.45VVITLGSPSDD583 pKa = 3.56QMILVGIQLSSLTPGWIFGAA603 pKa = 4.2
Molecular weight: 61.67 kDa
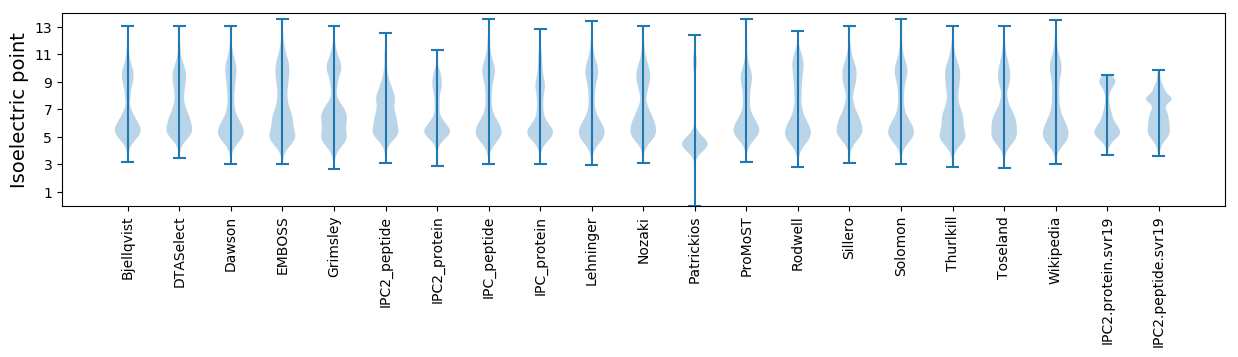
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A328BM32|A0A328BM32_9CAUL TSPc domain-containing protein OS=Phenylobacterium kunshanense OX=1445034 GN=DJ019_07480 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.47GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTKK25 pKa = 10.02NGQKK29 pKa = 9.43ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.47GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTKK25 pKa = 10.02NGQKK29 pKa = 9.43ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1261893 |
29 |
1949 |
311.8 |
33.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.127 ± 0.057 | 0.78 ± 0.013 |
5.811 ± 0.029 | 5.785 ± 0.035 |
3.502 ± 0.023 | 9.097 ± 0.04 |
1.809 ± 0.018 | 4.145 ± 0.027 |
2.983 ± 0.033 | 9.882 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.222 ± 0.018 | 2.226 ± 0.028 |
5.655 ± 0.032 | 3.048 ± 0.023 |
7.797 ± 0.043 | 4.819 ± 0.029 |
5.103 ± 0.031 | 7.642 ± 0.028 |
1.435 ± 0.015 | 2.133 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
