
Erigeron breviscapus amalgavirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; unclassified Amalgaviridae
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
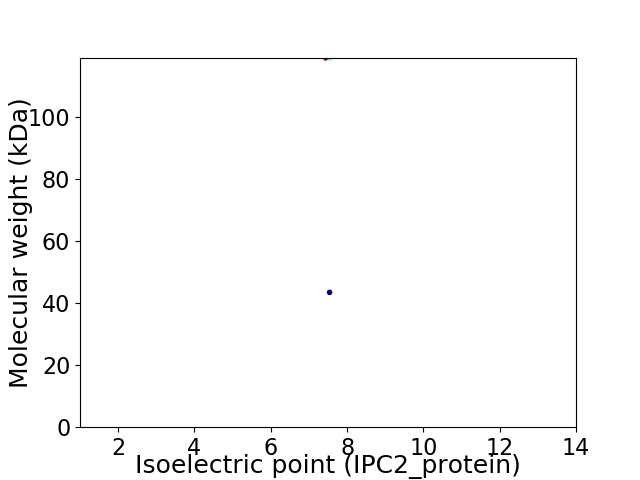
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z6JIJ9|A0A2Z6JIJ9_9VIRU ORF1+2p OS=Erigeron breviscapus amalgavirus 1 OX=2069322 GN=ORF1+ORF2 PE=4 SV=1
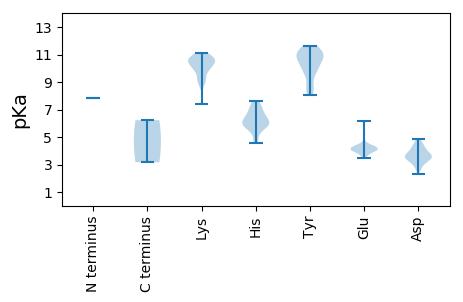
MM1 pKa = 7.81GEE3 pKa = 4.42PLPEE7 pKa = 3.97LTYY10 pKa = 11.22VLTPAEE16 pKa = 4.18EE17 pKa = 4.13QAEE20 pKa = 4.62LTRR23 pKa = 11.84LAAPLIQQGLPAGLFDD39 pKa = 3.5RR40 pKa = 11.84TAVLRR45 pKa = 11.84ANYY48 pKa = 8.59TYY50 pKa = 11.04KK51 pKa = 11.0GFLKK55 pKa = 10.52HH56 pKa = 5.84LQTVSRR62 pKa = 11.84LVDD65 pKa = 3.03QDD67 pKa = 4.1IIVDD71 pKa = 3.86ALSLGVKK78 pKa = 9.79KK79 pKa = 10.74DD80 pKa = 3.7FFPLPNRR87 pKa = 11.84MNISQFCRR95 pKa = 11.84FSEE98 pKa = 4.01WLRR101 pKa = 11.84SKK103 pKa = 10.87DD104 pKa = 3.66GQNSLHH110 pKa = 5.9EE111 pKa = 4.34VQRR114 pKa = 11.84HH115 pKa = 4.54KK116 pKa = 11.05KK117 pKa = 10.09LEE119 pKa = 4.11KK120 pKa = 9.93KK121 pKa = 10.28AAGVLEE127 pKa = 4.1PRR129 pKa = 11.84EE130 pKa = 4.03VALEE134 pKa = 4.08QIFSAQRR141 pKa = 11.84ADD143 pKa = 2.74WAAAKK148 pKa = 10.36KK149 pKa = 9.07EE150 pKa = 3.98EE151 pKa = 3.85RR152 pKa = 11.84SAYY155 pKa = 10.16DD156 pKa = 3.8KK157 pKa = 10.77EE158 pKa = 4.16IQEE161 pKa = 4.1LRR163 pKa = 11.84KK164 pKa = 10.02KK165 pKa = 10.05IRR167 pKa = 11.84QLEE170 pKa = 4.28RR171 pKa = 11.84RR172 pKa = 11.84WEE174 pKa = 3.97RR175 pKa = 11.84RR176 pKa = 11.84EE177 pKa = 3.88VEE179 pKa = 3.71IDD181 pKa = 3.28NAFDD185 pKa = 3.58PQFEE189 pKa = 4.09FLEE192 pKa = 4.37LDD194 pKa = 3.36EE195 pKa = 5.24AALNEE200 pKa = 4.23RR201 pKa = 11.84AYY203 pKa = 11.06AMYY206 pKa = 11.01VHH208 pKa = 7.48DD209 pKa = 4.52CQVKK213 pKa = 8.77EE214 pKa = 3.68RR215 pKa = 11.84RR216 pKa = 11.84PRR218 pKa = 11.84SKK220 pKa = 10.58EE221 pKa = 3.51HH222 pKa = 6.67GGVQLAVEE230 pKa = 4.28AFGGQVKK237 pKa = 9.76KK238 pKa = 10.73QKK240 pKa = 9.2QAEE243 pKa = 4.33FARR246 pKa = 11.84EE247 pKa = 3.69PEE249 pKa = 4.25VAVKK253 pKa = 10.48LFEE256 pKa = 3.97YY257 pKa = 10.35AKK259 pKa = 10.47RR260 pKa = 11.84KK261 pKa = 9.19ILSFEE266 pKa = 4.05EE267 pKa = 4.43MVTSNRR273 pKa = 11.84KK274 pKa = 9.08PIGKK278 pKa = 7.4FTWMPRR284 pKa = 11.84VEE286 pKa = 4.49TYY288 pKa = 10.97LLSRR292 pKa = 11.84PVKK295 pKa = 9.5LRR297 pKa = 11.84KK298 pKa = 9.49RR299 pKa = 11.84ICSMCPVGLPPPPGVAPGCQPLSKK323 pKa = 10.25FVRR326 pKa = 11.84EE327 pKa = 4.0EE328 pKa = 3.94LLVSGVVEE336 pKa = 4.49GTKK339 pKa = 8.78KK340 pKa = 9.35TNPLLRR346 pKa = 11.84AFATVQLGAEE356 pKa = 4.53VLRR359 pKa = 11.84STRR362 pKa = 11.84VLVHH366 pKa = 5.67HH367 pKa = 6.55TFGPEE372 pKa = 3.59TQRR375 pKa = 11.84RR376 pKa = 11.84IPVARR381 pKa = 11.84SRR383 pKa = 11.84IEE385 pKa = 3.66AGLRR389 pKa = 11.84RR390 pKa = 11.84IIGGGAMRR398 pKa = 11.84GWDD401 pKa = 3.79ADD403 pKa = 3.31SKK405 pKa = 9.87MFRR408 pKa = 11.84GGGNSSDD415 pKa = 3.7ALLLLGQCDD424 pKa = 3.94DD425 pKa = 4.34NLPGGLLRR433 pKa = 11.84EE434 pKa = 4.32HH435 pKa = 7.1FSLLSAKK442 pKa = 10.02RR443 pKa = 11.84ALSLPGGLRR452 pKa = 11.84VPDD455 pKa = 4.22GPEE458 pKa = 3.76CLVMKK463 pKa = 10.39NFNNDD468 pKa = 2.34ATAGPFLRR476 pKa = 11.84AFGIKK481 pKa = 9.77GKK483 pKa = 10.85YY484 pKa = 8.38GLKK487 pKa = 10.36KK488 pKa = 10.53LLEE491 pKa = 4.23DD492 pKa = 3.92TMWWFYY498 pKa = 11.32DD499 pKa = 3.19AYY501 pKa = 10.53GRR503 pKa = 11.84GEE505 pKa = 3.96ISDD508 pKa = 4.17GEE510 pKa = 4.38MPHH513 pKa = 6.08FAARR517 pKa = 11.84VGFRR521 pKa = 11.84TKK523 pKa = 10.53LVSEE527 pKa = 4.57TKK529 pKa = 10.29AWEE532 pKa = 4.04KK533 pKa = 10.44LAAGAPVGRR542 pKa = 11.84AVMMLDD548 pKa = 3.68ALEE551 pKa = 4.23QAASSPLYY559 pKa = 10.78NVMSNSTYY567 pKa = 8.49QRR569 pKa = 11.84RR570 pKa = 11.84LEE572 pKa = 4.14RR573 pKa = 11.84DD574 pKa = 3.29CGFKK578 pKa = 10.82NGIVKK583 pKa = 10.64ASSDD587 pKa = 3.09WAKK590 pKa = 9.4IWEE593 pKa = 4.35DD594 pKa = 3.26VRR596 pKa = 11.84EE597 pKa = 4.15AKK599 pKa = 10.51AIIEE603 pKa = 4.2LDD605 pKa = 2.79WSKK608 pKa = 10.98FDD610 pKa = 4.39RR611 pKa = 11.84EE612 pKa = 4.13RR613 pKa = 11.84PADD616 pKa = 4.63DD617 pKa = 3.13ILFVIEE623 pKa = 4.31VVLSCFEE630 pKa = 3.96PTNDD634 pKa = 3.58RR635 pKa = 11.84EE636 pKa = 4.18RR637 pKa = 11.84RR638 pKa = 11.84LLRR641 pKa = 11.84AFGLMMRR648 pKa = 11.84RR649 pKa = 11.84ALVEE653 pKa = 4.31RR654 pKa = 11.84IIVMDD659 pKa = 4.28DD660 pKa = 3.26GGVFEE665 pKa = 5.25IDD667 pKa = 3.19GMVPSGSLWTGWLDD681 pKa = 3.27TALNVLYY688 pKa = 11.01LNAACMNVGIGPLGFSAMCAGDD710 pKa = 4.73DD711 pKa = 3.97NLTLFWTDD719 pKa = 3.5HH720 pKa = 6.94PDD722 pKa = 3.38HH723 pKa = 6.08VLKK726 pKa = 10.74RR727 pKa = 11.84IKK729 pKa = 10.69DD730 pKa = 3.6EE731 pKa = 4.35LNGKK735 pKa = 8.76FRR737 pKa = 11.84AGISDD742 pKa = 3.56EE743 pKa = 4.88DD744 pKa = 4.33FFIHH748 pKa = 7.07RR749 pKa = 11.84PPFHH753 pKa = 5.87VTKK756 pKa = 10.44QQACFPPGTDD766 pKa = 3.76LSHH769 pKa = 6.04GTSKK773 pKa = 11.07LMDD776 pKa = 3.6LVFWQVFDD784 pKa = 4.12GEE786 pKa = 4.69VVIDD790 pKa = 3.88EE791 pKa = 4.36AAGRR795 pKa = 11.84SHH797 pKa = 6.4RR798 pKa = 11.84WEE800 pKa = 3.97YY801 pKa = 10.42VFKK804 pKa = 10.79GRR806 pKa = 11.84PKK808 pKa = 10.44FLSCYY813 pKa = 8.04WLPGGQPIRR822 pKa = 11.84PTSDD826 pKa = 2.72NMEE829 pKa = 4.22KK830 pKa = 10.73LLWPEE835 pKa = 5.26GIHH838 pKa = 7.04EE839 pKa = 6.15DD840 pKa = 3.61IDD842 pKa = 4.85DD843 pKa = 4.04YY844 pKa = 11.5QATVMAMVVDD854 pKa = 4.36NPWNHH859 pKa = 6.23HH860 pKa = 5.61CVNHH864 pKa = 6.19LLMRR868 pKa = 11.84YY869 pKa = 9.63VILQQLRR876 pKa = 11.84RR877 pKa = 11.84VDD879 pKa = 3.43ILRR882 pKa = 11.84GGMDD886 pKa = 4.48DD887 pKa = 4.85VLFLCACRR895 pKa = 11.84EE896 pKa = 4.13KK897 pKa = 11.14GGGPIPYY904 pKa = 9.81PMVAPWRR911 pKa = 11.84RR912 pKa = 11.84SEE914 pKa = 3.69VHH916 pKa = 6.38GRR918 pKa = 11.84MEE920 pKa = 4.99DD921 pKa = 3.56YY922 pKa = 11.33EE923 pKa = 4.37EE924 pKa = 4.28VKK926 pKa = 10.71RR927 pKa = 11.84HH928 pKa = 5.44IQDD931 pKa = 3.08FSDD934 pKa = 3.71FVTGVTSLYY943 pKa = 10.89SRR945 pKa = 11.84TATGGVDD952 pKa = 2.57SWLFMNIIRR961 pKa = 11.84GEE963 pKa = 3.96QHH965 pKa = 6.07VGEE968 pKa = 4.3GQYY971 pKa = 11.66GNDD974 pKa = 3.07LMVWVSWIRR983 pKa = 11.84DD984 pKa = 3.89HH985 pKa = 7.6PCTRR989 pKa = 11.84YY990 pKa = 9.67LKK992 pKa = 10.02SVRR995 pKa = 11.84GLRR998 pKa = 11.84TRR1000 pKa = 11.84VEE1002 pKa = 4.07QLDD1005 pKa = 3.79VDD1007 pKa = 4.1PGLLQKK1013 pKa = 10.2ATIHH1017 pKa = 5.94YY1018 pKa = 9.58SLLRR1022 pKa = 11.84EE1023 pKa = 4.07TLVSGRR1029 pKa = 11.84IEE1031 pKa = 3.99TALDD1035 pKa = 3.46FARR1038 pKa = 11.84WVRR1041 pKa = 11.84SIILGGHH1048 pKa = 4.98VV1049 pKa = 3.17
MM1 pKa = 7.81GEE3 pKa = 4.42PLPEE7 pKa = 3.97LTYY10 pKa = 11.22VLTPAEE16 pKa = 4.18EE17 pKa = 4.13QAEE20 pKa = 4.62LTRR23 pKa = 11.84LAAPLIQQGLPAGLFDD39 pKa = 3.5RR40 pKa = 11.84TAVLRR45 pKa = 11.84ANYY48 pKa = 8.59TYY50 pKa = 11.04KK51 pKa = 11.0GFLKK55 pKa = 10.52HH56 pKa = 5.84LQTVSRR62 pKa = 11.84LVDD65 pKa = 3.03QDD67 pKa = 4.1IIVDD71 pKa = 3.86ALSLGVKK78 pKa = 9.79KK79 pKa = 10.74DD80 pKa = 3.7FFPLPNRR87 pKa = 11.84MNISQFCRR95 pKa = 11.84FSEE98 pKa = 4.01WLRR101 pKa = 11.84SKK103 pKa = 10.87DD104 pKa = 3.66GQNSLHH110 pKa = 5.9EE111 pKa = 4.34VQRR114 pKa = 11.84HH115 pKa = 4.54KK116 pKa = 11.05KK117 pKa = 10.09LEE119 pKa = 4.11KK120 pKa = 9.93KK121 pKa = 10.28AAGVLEE127 pKa = 4.1PRR129 pKa = 11.84EE130 pKa = 4.03VALEE134 pKa = 4.08QIFSAQRR141 pKa = 11.84ADD143 pKa = 2.74WAAAKK148 pKa = 10.36KK149 pKa = 9.07EE150 pKa = 3.98EE151 pKa = 3.85RR152 pKa = 11.84SAYY155 pKa = 10.16DD156 pKa = 3.8KK157 pKa = 10.77EE158 pKa = 4.16IQEE161 pKa = 4.1LRR163 pKa = 11.84KK164 pKa = 10.02KK165 pKa = 10.05IRR167 pKa = 11.84QLEE170 pKa = 4.28RR171 pKa = 11.84RR172 pKa = 11.84WEE174 pKa = 3.97RR175 pKa = 11.84RR176 pKa = 11.84EE177 pKa = 3.88VEE179 pKa = 3.71IDD181 pKa = 3.28NAFDD185 pKa = 3.58PQFEE189 pKa = 4.09FLEE192 pKa = 4.37LDD194 pKa = 3.36EE195 pKa = 5.24AALNEE200 pKa = 4.23RR201 pKa = 11.84AYY203 pKa = 11.06AMYY206 pKa = 11.01VHH208 pKa = 7.48DD209 pKa = 4.52CQVKK213 pKa = 8.77EE214 pKa = 3.68RR215 pKa = 11.84RR216 pKa = 11.84PRR218 pKa = 11.84SKK220 pKa = 10.58EE221 pKa = 3.51HH222 pKa = 6.67GGVQLAVEE230 pKa = 4.28AFGGQVKK237 pKa = 9.76KK238 pKa = 10.73QKK240 pKa = 9.2QAEE243 pKa = 4.33FARR246 pKa = 11.84EE247 pKa = 3.69PEE249 pKa = 4.25VAVKK253 pKa = 10.48LFEE256 pKa = 3.97YY257 pKa = 10.35AKK259 pKa = 10.47RR260 pKa = 11.84KK261 pKa = 9.19ILSFEE266 pKa = 4.05EE267 pKa = 4.43MVTSNRR273 pKa = 11.84KK274 pKa = 9.08PIGKK278 pKa = 7.4FTWMPRR284 pKa = 11.84VEE286 pKa = 4.49TYY288 pKa = 10.97LLSRR292 pKa = 11.84PVKK295 pKa = 9.5LRR297 pKa = 11.84KK298 pKa = 9.49RR299 pKa = 11.84ICSMCPVGLPPPPGVAPGCQPLSKK323 pKa = 10.25FVRR326 pKa = 11.84EE327 pKa = 4.0EE328 pKa = 3.94LLVSGVVEE336 pKa = 4.49GTKK339 pKa = 8.78KK340 pKa = 9.35TNPLLRR346 pKa = 11.84AFATVQLGAEE356 pKa = 4.53VLRR359 pKa = 11.84STRR362 pKa = 11.84VLVHH366 pKa = 5.67HH367 pKa = 6.55TFGPEE372 pKa = 3.59TQRR375 pKa = 11.84RR376 pKa = 11.84IPVARR381 pKa = 11.84SRR383 pKa = 11.84IEE385 pKa = 3.66AGLRR389 pKa = 11.84RR390 pKa = 11.84IIGGGAMRR398 pKa = 11.84GWDD401 pKa = 3.79ADD403 pKa = 3.31SKK405 pKa = 9.87MFRR408 pKa = 11.84GGGNSSDD415 pKa = 3.7ALLLLGQCDD424 pKa = 3.94DD425 pKa = 4.34NLPGGLLRR433 pKa = 11.84EE434 pKa = 4.32HH435 pKa = 7.1FSLLSAKK442 pKa = 10.02RR443 pKa = 11.84ALSLPGGLRR452 pKa = 11.84VPDD455 pKa = 4.22GPEE458 pKa = 3.76CLVMKK463 pKa = 10.39NFNNDD468 pKa = 2.34ATAGPFLRR476 pKa = 11.84AFGIKK481 pKa = 9.77GKK483 pKa = 10.85YY484 pKa = 8.38GLKK487 pKa = 10.36KK488 pKa = 10.53LLEE491 pKa = 4.23DD492 pKa = 3.92TMWWFYY498 pKa = 11.32DD499 pKa = 3.19AYY501 pKa = 10.53GRR503 pKa = 11.84GEE505 pKa = 3.96ISDD508 pKa = 4.17GEE510 pKa = 4.38MPHH513 pKa = 6.08FAARR517 pKa = 11.84VGFRR521 pKa = 11.84TKK523 pKa = 10.53LVSEE527 pKa = 4.57TKK529 pKa = 10.29AWEE532 pKa = 4.04KK533 pKa = 10.44LAAGAPVGRR542 pKa = 11.84AVMMLDD548 pKa = 3.68ALEE551 pKa = 4.23QAASSPLYY559 pKa = 10.78NVMSNSTYY567 pKa = 8.49QRR569 pKa = 11.84RR570 pKa = 11.84LEE572 pKa = 4.14RR573 pKa = 11.84DD574 pKa = 3.29CGFKK578 pKa = 10.82NGIVKK583 pKa = 10.64ASSDD587 pKa = 3.09WAKK590 pKa = 9.4IWEE593 pKa = 4.35DD594 pKa = 3.26VRR596 pKa = 11.84EE597 pKa = 4.15AKK599 pKa = 10.51AIIEE603 pKa = 4.2LDD605 pKa = 2.79WSKK608 pKa = 10.98FDD610 pKa = 4.39RR611 pKa = 11.84EE612 pKa = 4.13RR613 pKa = 11.84PADD616 pKa = 4.63DD617 pKa = 3.13ILFVIEE623 pKa = 4.31VVLSCFEE630 pKa = 3.96PTNDD634 pKa = 3.58RR635 pKa = 11.84EE636 pKa = 4.18RR637 pKa = 11.84RR638 pKa = 11.84LLRR641 pKa = 11.84AFGLMMRR648 pKa = 11.84RR649 pKa = 11.84ALVEE653 pKa = 4.31RR654 pKa = 11.84IIVMDD659 pKa = 4.28DD660 pKa = 3.26GGVFEE665 pKa = 5.25IDD667 pKa = 3.19GMVPSGSLWTGWLDD681 pKa = 3.27TALNVLYY688 pKa = 11.01LNAACMNVGIGPLGFSAMCAGDD710 pKa = 4.73DD711 pKa = 3.97NLTLFWTDD719 pKa = 3.5HH720 pKa = 6.94PDD722 pKa = 3.38HH723 pKa = 6.08VLKK726 pKa = 10.74RR727 pKa = 11.84IKK729 pKa = 10.69DD730 pKa = 3.6EE731 pKa = 4.35LNGKK735 pKa = 8.76FRR737 pKa = 11.84AGISDD742 pKa = 3.56EE743 pKa = 4.88DD744 pKa = 4.33FFIHH748 pKa = 7.07RR749 pKa = 11.84PPFHH753 pKa = 5.87VTKK756 pKa = 10.44QQACFPPGTDD766 pKa = 3.76LSHH769 pKa = 6.04GTSKK773 pKa = 11.07LMDD776 pKa = 3.6LVFWQVFDD784 pKa = 4.12GEE786 pKa = 4.69VVIDD790 pKa = 3.88EE791 pKa = 4.36AAGRR795 pKa = 11.84SHH797 pKa = 6.4RR798 pKa = 11.84WEE800 pKa = 3.97YY801 pKa = 10.42VFKK804 pKa = 10.79GRR806 pKa = 11.84PKK808 pKa = 10.44FLSCYY813 pKa = 8.04WLPGGQPIRR822 pKa = 11.84PTSDD826 pKa = 2.72NMEE829 pKa = 4.22KK830 pKa = 10.73LLWPEE835 pKa = 5.26GIHH838 pKa = 7.04EE839 pKa = 6.15DD840 pKa = 3.61IDD842 pKa = 4.85DD843 pKa = 4.04YY844 pKa = 11.5QATVMAMVVDD854 pKa = 4.36NPWNHH859 pKa = 6.23HH860 pKa = 5.61CVNHH864 pKa = 6.19LLMRR868 pKa = 11.84YY869 pKa = 9.63VILQQLRR876 pKa = 11.84RR877 pKa = 11.84VDD879 pKa = 3.43ILRR882 pKa = 11.84GGMDD886 pKa = 4.48DD887 pKa = 4.85VLFLCACRR895 pKa = 11.84EE896 pKa = 4.13KK897 pKa = 11.14GGGPIPYY904 pKa = 9.81PMVAPWRR911 pKa = 11.84RR912 pKa = 11.84SEE914 pKa = 3.69VHH916 pKa = 6.38GRR918 pKa = 11.84MEE920 pKa = 4.99DD921 pKa = 3.56YY922 pKa = 11.33EE923 pKa = 4.37EE924 pKa = 4.28VKK926 pKa = 10.71RR927 pKa = 11.84HH928 pKa = 5.44IQDD931 pKa = 3.08FSDD934 pKa = 3.71FVTGVTSLYY943 pKa = 10.89SRR945 pKa = 11.84TATGGVDD952 pKa = 2.57SWLFMNIIRR961 pKa = 11.84GEE963 pKa = 3.96QHH965 pKa = 6.07VGEE968 pKa = 4.3GQYY971 pKa = 11.66GNDD974 pKa = 3.07LMVWVSWIRR983 pKa = 11.84DD984 pKa = 3.89HH985 pKa = 7.6PCTRR989 pKa = 11.84YY990 pKa = 9.67LKK992 pKa = 10.02SVRR995 pKa = 11.84GLRR998 pKa = 11.84TRR1000 pKa = 11.84VEE1002 pKa = 4.07QLDD1005 pKa = 3.79VDD1007 pKa = 4.1PGLLQKK1013 pKa = 10.2ATIHH1017 pKa = 5.94YY1018 pKa = 9.58SLLRR1022 pKa = 11.84EE1023 pKa = 4.07TLVSGRR1029 pKa = 11.84IEE1031 pKa = 3.99TALDD1035 pKa = 3.46FARR1038 pKa = 11.84WVRR1041 pKa = 11.84SIILGGHH1048 pKa = 4.98VV1049 pKa = 3.17
Molecular weight: 119.12 kDa
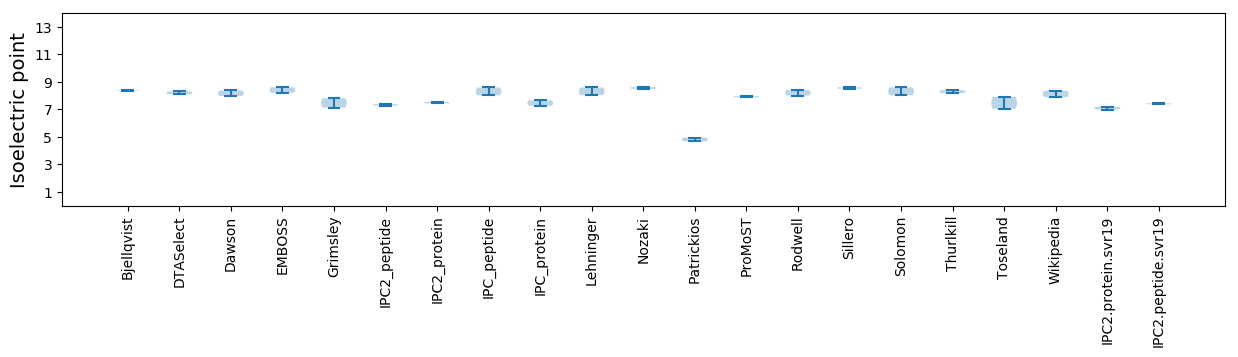
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z6JIJ9|A0A2Z6JIJ9_9VIRU ORF1+2p OS=Erigeron breviscapus amalgavirus 1 OX=2069322 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.81GEE3 pKa = 4.42PLPEE7 pKa = 3.97LTYY10 pKa = 11.22VLTPAEE16 pKa = 4.18EE17 pKa = 4.13QAEE20 pKa = 4.62LTRR23 pKa = 11.84LAAPLIQQGLPAGLFDD39 pKa = 3.5RR40 pKa = 11.84TAVLRR45 pKa = 11.84ANYY48 pKa = 8.59TYY50 pKa = 11.04KK51 pKa = 11.0GFLKK55 pKa = 10.52HH56 pKa = 5.84LQTVSRR62 pKa = 11.84LVDD65 pKa = 3.03QDD67 pKa = 4.1IIVDD71 pKa = 3.86ALSLGVKK78 pKa = 9.79KK79 pKa = 10.74DD80 pKa = 3.7FFPLPNRR87 pKa = 11.84MNISQFCRR95 pKa = 11.84FSEE98 pKa = 4.01WLRR101 pKa = 11.84SKK103 pKa = 10.87DD104 pKa = 3.66GQNSLHH110 pKa = 5.9EE111 pKa = 4.34VQRR114 pKa = 11.84HH115 pKa = 4.54KK116 pKa = 11.05KK117 pKa = 10.09LEE119 pKa = 4.11KK120 pKa = 9.93KK121 pKa = 10.28AAGVLEE127 pKa = 4.1PRR129 pKa = 11.84EE130 pKa = 4.03VALEE134 pKa = 4.08QIFSAQRR141 pKa = 11.84ADD143 pKa = 2.74WAAAKK148 pKa = 10.36KK149 pKa = 9.07EE150 pKa = 3.98EE151 pKa = 3.85RR152 pKa = 11.84SAYY155 pKa = 10.16DD156 pKa = 3.8KK157 pKa = 10.77EE158 pKa = 4.16IQEE161 pKa = 4.1LRR163 pKa = 11.84KK164 pKa = 10.02KK165 pKa = 10.05IRR167 pKa = 11.84QLEE170 pKa = 4.28RR171 pKa = 11.84RR172 pKa = 11.84WEE174 pKa = 3.97RR175 pKa = 11.84RR176 pKa = 11.84EE177 pKa = 3.88VEE179 pKa = 3.71IDD181 pKa = 3.28NAFDD185 pKa = 3.58PQFEE189 pKa = 4.09FLEE192 pKa = 4.37LDD194 pKa = 3.36EE195 pKa = 5.24AALNEE200 pKa = 4.23RR201 pKa = 11.84AYY203 pKa = 11.06AMYY206 pKa = 11.01VHH208 pKa = 7.48DD209 pKa = 4.52CQVKK213 pKa = 8.77EE214 pKa = 3.68RR215 pKa = 11.84RR216 pKa = 11.84PRR218 pKa = 11.84SKK220 pKa = 10.58EE221 pKa = 3.51HH222 pKa = 6.67GGVQLAVEE230 pKa = 4.28AFGGQVKK237 pKa = 9.76KK238 pKa = 10.73QKK240 pKa = 9.2QAEE243 pKa = 4.33FARR246 pKa = 11.84EE247 pKa = 3.69PEE249 pKa = 4.25VAVKK253 pKa = 10.48LFEE256 pKa = 3.97YY257 pKa = 10.35AKK259 pKa = 10.52RR260 pKa = 11.84KK261 pKa = 9.19ILSFRR266 pKa = 11.84RR267 pKa = 11.84DD268 pKa = 2.84GDD270 pKa = 3.5IKK272 pKa = 11.08QEE274 pKa = 4.31ANWKK278 pKa = 10.16VYY280 pKa = 9.84MDD282 pKa = 4.07AAGGDD287 pKa = 4.22LSTVPTGEE295 pKa = 3.99APEE298 pKa = 4.34EE299 pKa = 3.86NLLYY303 pKa = 10.45VPRR306 pKa = 11.84GVTPPPRR313 pKa = 11.84GGPGVPTAQQVRR325 pKa = 11.84QGGVTGVRR333 pKa = 11.84RR334 pKa = 11.84GRR336 pKa = 11.84GDD338 pKa = 4.61EE339 pKa = 3.97EE340 pKa = 4.29DD341 pKa = 3.52QPIAARR347 pKa = 11.84LRR349 pKa = 11.84NRR351 pKa = 11.84PTRR354 pKa = 11.84SRR356 pKa = 11.84SIEE359 pKa = 3.77EE360 pKa = 3.93HH361 pKa = 5.85QGVGTSHH368 pKa = 6.61IRR370 pKa = 11.84AGDD373 pKa = 3.58TTEE376 pKa = 4.35DD377 pKa = 3.42TGSSLPHH384 pKa = 6.23
MM1 pKa = 7.81GEE3 pKa = 4.42PLPEE7 pKa = 3.97LTYY10 pKa = 11.22VLTPAEE16 pKa = 4.18EE17 pKa = 4.13QAEE20 pKa = 4.62LTRR23 pKa = 11.84LAAPLIQQGLPAGLFDD39 pKa = 3.5RR40 pKa = 11.84TAVLRR45 pKa = 11.84ANYY48 pKa = 8.59TYY50 pKa = 11.04KK51 pKa = 11.0GFLKK55 pKa = 10.52HH56 pKa = 5.84LQTVSRR62 pKa = 11.84LVDD65 pKa = 3.03QDD67 pKa = 4.1IIVDD71 pKa = 3.86ALSLGVKK78 pKa = 9.79KK79 pKa = 10.74DD80 pKa = 3.7FFPLPNRR87 pKa = 11.84MNISQFCRR95 pKa = 11.84FSEE98 pKa = 4.01WLRR101 pKa = 11.84SKK103 pKa = 10.87DD104 pKa = 3.66GQNSLHH110 pKa = 5.9EE111 pKa = 4.34VQRR114 pKa = 11.84HH115 pKa = 4.54KK116 pKa = 11.05KK117 pKa = 10.09LEE119 pKa = 4.11KK120 pKa = 9.93KK121 pKa = 10.28AAGVLEE127 pKa = 4.1PRR129 pKa = 11.84EE130 pKa = 4.03VALEE134 pKa = 4.08QIFSAQRR141 pKa = 11.84ADD143 pKa = 2.74WAAAKK148 pKa = 10.36KK149 pKa = 9.07EE150 pKa = 3.98EE151 pKa = 3.85RR152 pKa = 11.84SAYY155 pKa = 10.16DD156 pKa = 3.8KK157 pKa = 10.77EE158 pKa = 4.16IQEE161 pKa = 4.1LRR163 pKa = 11.84KK164 pKa = 10.02KK165 pKa = 10.05IRR167 pKa = 11.84QLEE170 pKa = 4.28RR171 pKa = 11.84RR172 pKa = 11.84WEE174 pKa = 3.97RR175 pKa = 11.84RR176 pKa = 11.84EE177 pKa = 3.88VEE179 pKa = 3.71IDD181 pKa = 3.28NAFDD185 pKa = 3.58PQFEE189 pKa = 4.09FLEE192 pKa = 4.37LDD194 pKa = 3.36EE195 pKa = 5.24AALNEE200 pKa = 4.23RR201 pKa = 11.84AYY203 pKa = 11.06AMYY206 pKa = 11.01VHH208 pKa = 7.48DD209 pKa = 4.52CQVKK213 pKa = 8.77EE214 pKa = 3.68RR215 pKa = 11.84RR216 pKa = 11.84PRR218 pKa = 11.84SKK220 pKa = 10.58EE221 pKa = 3.51HH222 pKa = 6.67GGVQLAVEE230 pKa = 4.28AFGGQVKK237 pKa = 9.76KK238 pKa = 10.73QKK240 pKa = 9.2QAEE243 pKa = 4.33FARR246 pKa = 11.84EE247 pKa = 3.69PEE249 pKa = 4.25VAVKK253 pKa = 10.48LFEE256 pKa = 3.97YY257 pKa = 10.35AKK259 pKa = 10.52RR260 pKa = 11.84KK261 pKa = 9.19ILSFRR266 pKa = 11.84RR267 pKa = 11.84DD268 pKa = 2.84GDD270 pKa = 3.5IKK272 pKa = 11.08QEE274 pKa = 4.31ANWKK278 pKa = 10.16VYY280 pKa = 9.84MDD282 pKa = 4.07AAGGDD287 pKa = 4.22LSTVPTGEE295 pKa = 3.99APEE298 pKa = 4.34EE299 pKa = 3.86NLLYY303 pKa = 10.45VPRR306 pKa = 11.84GVTPPPRR313 pKa = 11.84GGPGVPTAQQVRR325 pKa = 11.84QGGVTGVRR333 pKa = 11.84RR334 pKa = 11.84GRR336 pKa = 11.84GDD338 pKa = 4.61EE339 pKa = 3.97EE340 pKa = 4.29DD341 pKa = 3.52QPIAARR347 pKa = 11.84LRR349 pKa = 11.84NRR351 pKa = 11.84PTRR354 pKa = 11.84SRR356 pKa = 11.84SIEE359 pKa = 3.77EE360 pKa = 3.93HH361 pKa = 5.85QGVGTSHH368 pKa = 6.61IRR370 pKa = 11.84AGDD373 pKa = 3.58TTEE376 pKa = 4.35DD377 pKa = 3.42TGSSLPHH384 pKa = 6.23
Molecular weight: 43.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1433 |
384 |
1049 |
716.5 |
81.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.886 ± 0.775 | 1.326 ± 0.419 |
5.792 ± 0.304 | 7.955 ± 1.145 |
4.327 ± 0.354 | 7.746 ± 0.236 |
2.303 ± 0.114 | 4.047 ± 0.344 |
5.583 ± 0.347 | 9.839 ± 0.648 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.373 ± 0.692 | 2.582 ± 0.124 |
5.234 ± 0.258 | 4.187 ± 1.073 |
8.723 ± 0.475 | 4.745 ± 0.301 |
3.838 ± 0.171 | 7.258 ± 0.253 |
1.954 ± 0.475 | 2.303 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
