
Candidatus Viridilinea mediisalina
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Chloroflexia; Chloroflexales; Chloroflexineae; Oscillochloridaceae; Candidatus Viridilinea
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
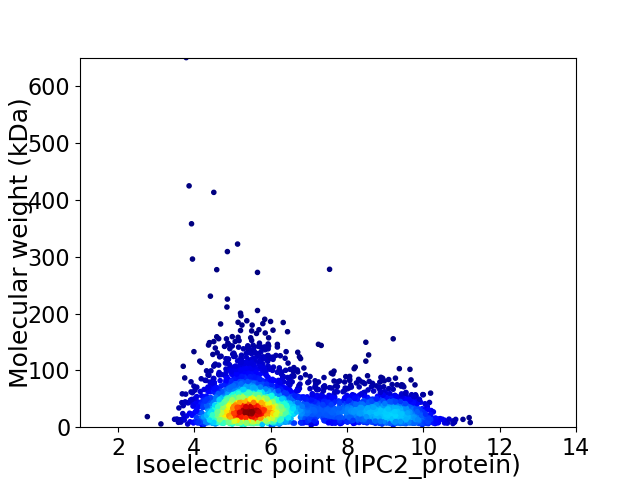
Virtual 2D-PAGE plot for 4287 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A6RGH5|A0A2A6RGH5_9CHLR NHase_alpha domain-containing protein OS=Candidatus Viridilinea mediisalina OX=2024553 GN=CJ255_16240 PE=4 SV=1
MM1 pKa = 7.51LAACGGGTQPAAAPPAPPAEE21 pKa = 4.67AEE23 pKa = 4.59DD24 pKa = 3.86VTEE27 pKa = 5.36PDD29 pKa = 3.28ATDD32 pKa = 3.74DD33 pKa = 5.47AEE35 pKa = 4.39ATDD38 pKa = 4.42DD39 pKa = 5.76AEE41 pKa = 4.52ATDD44 pKa = 4.73DD45 pKa = 5.45ADD47 pKa = 3.76VTDD50 pKa = 4.88DD51 pKa = 4.24AEE53 pKa = 4.48EE54 pKa = 4.16PAAPADD60 pKa = 3.98PADD63 pKa = 4.01GPMLYY68 pKa = 10.15YY69 pKa = 10.84AFGGGPVGGVFQVYY83 pKa = 10.43AEE85 pKa = 4.18ALSLIINAAVPNVDD99 pKa = 3.66LTAEE103 pKa = 4.33GTGGSAEE110 pKa = 4.12NLRR113 pKa = 11.84SVNSGDD119 pKa = 3.42RR120 pKa = 11.84QFGIVYY126 pKa = 10.33AGDD129 pKa = 3.33VSLGAEE135 pKa = 4.04GLLPEE140 pKa = 5.71DD141 pKa = 3.9ANQYY145 pKa = 8.16TNVMPVAPLYY155 pKa = 10.95GGVIHH160 pKa = 6.67LVVSRR165 pKa = 11.84ASGIEE170 pKa = 3.99TVADD174 pKa = 3.82LPGKK178 pKa = 10.47RR179 pKa = 11.84IALGNAGSGAALSAEE194 pKa = 4.14RR195 pKa = 11.84YY196 pKa = 7.44FNHH199 pKa = 6.64MGLMDD204 pKa = 4.01QVNVEE209 pKa = 4.16YY210 pKa = 10.74LGYY213 pKa = 9.47SQAAAAMGDD222 pKa = 3.75GQLDD226 pKa = 4.11GFWILAGFPNASVTEE241 pKa = 4.08ASTYY245 pKa = 9.73TDD247 pKa = 2.43IRR249 pKa = 11.84LIDD252 pKa = 3.7VYY254 pKa = 11.49NPGVEE259 pKa = 4.12GGFFEE264 pKa = 6.01AYY266 pKa = 9.17PFYY269 pKa = 10.98SFRR272 pKa = 11.84ALDD275 pKa = 3.66GGAYY279 pKa = 9.23PGNPEE284 pKa = 4.74DD285 pKa = 3.96VPSFQDD291 pKa = 3.18TALWVAHH298 pKa = 6.23TDD300 pKa = 3.4VPEE303 pKa = 4.23DD304 pKa = 3.39VVYY307 pKa = 10.86NALRR311 pKa = 11.84AVFLDD316 pKa = 3.63GGLDD320 pKa = 3.23RR321 pKa = 11.84MVATHH326 pKa = 7.42ASAQEE331 pKa = 3.84MGLEE335 pKa = 3.99VGITGIANQLHH346 pKa = 6.65PGAVRR351 pKa = 11.84FWEE354 pKa = 4.33EE355 pKa = 3.65QGVTIPDD362 pKa = 3.54EE363 pKa = 4.14LRR365 pKa = 4.8
MM1 pKa = 7.51LAACGGGTQPAAAPPAPPAEE21 pKa = 4.67AEE23 pKa = 4.59DD24 pKa = 3.86VTEE27 pKa = 5.36PDD29 pKa = 3.28ATDD32 pKa = 3.74DD33 pKa = 5.47AEE35 pKa = 4.39ATDD38 pKa = 4.42DD39 pKa = 5.76AEE41 pKa = 4.52ATDD44 pKa = 4.73DD45 pKa = 5.45ADD47 pKa = 3.76VTDD50 pKa = 4.88DD51 pKa = 4.24AEE53 pKa = 4.48EE54 pKa = 4.16PAAPADD60 pKa = 3.98PADD63 pKa = 4.01GPMLYY68 pKa = 10.15YY69 pKa = 10.84AFGGGPVGGVFQVYY83 pKa = 10.43AEE85 pKa = 4.18ALSLIINAAVPNVDD99 pKa = 3.66LTAEE103 pKa = 4.33GTGGSAEE110 pKa = 4.12NLRR113 pKa = 11.84SVNSGDD119 pKa = 3.42RR120 pKa = 11.84QFGIVYY126 pKa = 10.33AGDD129 pKa = 3.33VSLGAEE135 pKa = 4.04GLLPEE140 pKa = 5.71DD141 pKa = 3.9ANQYY145 pKa = 8.16TNVMPVAPLYY155 pKa = 10.95GGVIHH160 pKa = 6.67LVVSRR165 pKa = 11.84ASGIEE170 pKa = 3.99TVADD174 pKa = 3.82LPGKK178 pKa = 10.47RR179 pKa = 11.84IALGNAGSGAALSAEE194 pKa = 4.14RR195 pKa = 11.84YY196 pKa = 7.44FNHH199 pKa = 6.64MGLMDD204 pKa = 4.01QVNVEE209 pKa = 4.16YY210 pKa = 10.74LGYY213 pKa = 9.47SQAAAAMGDD222 pKa = 3.75GQLDD226 pKa = 4.11GFWILAGFPNASVTEE241 pKa = 4.08ASTYY245 pKa = 9.73TDD247 pKa = 2.43IRR249 pKa = 11.84LIDD252 pKa = 3.7VYY254 pKa = 11.49NPGVEE259 pKa = 4.12GGFFEE264 pKa = 6.01AYY266 pKa = 9.17PFYY269 pKa = 10.98SFRR272 pKa = 11.84ALDD275 pKa = 3.66GGAYY279 pKa = 9.23PGNPEE284 pKa = 4.74DD285 pKa = 3.96VPSFQDD291 pKa = 3.18TALWVAHH298 pKa = 6.23TDD300 pKa = 3.4VPEE303 pKa = 4.23DD304 pKa = 3.39VVYY307 pKa = 10.86NALRR311 pKa = 11.84AVFLDD316 pKa = 3.63GGLDD320 pKa = 3.23RR321 pKa = 11.84MVATHH326 pKa = 7.42ASAQEE331 pKa = 3.84MGLEE335 pKa = 3.99VGITGIANQLHH346 pKa = 6.65PGAVRR351 pKa = 11.84FWEE354 pKa = 4.33EE355 pKa = 3.65QGVTIPDD362 pKa = 3.54EE363 pKa = 4.14LRR365 pKa = 4.8
Molecular weight: 37.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A6RJL5|A0A2A6RJL5_9CHLR Uncharacterized protein OS=Candidatus Viridilinea mediisalina OX=2024553 GN=CJ255_09795 PE=4 SV=1
MM1 pKa = 7.94RR2 pKa = 11.84SQHH5 pKa = 5.94ILASSRR11 pKa = 11.84PRR13 pKa = 11.84LLAPSPPRR21 pKa = 11.84ALASSPPRR29 pKa = 11.84LLASSPPRR37 pKa = 11.84LLASSPPRR45 pKa = 11.84LLASSPPRR53 pKa = 11.84LLASSPPRR61 pKa = 11.84LLASSPPRR69 pKa = 11.84LLASSPPRR77 pKa = 11.84LL78 pKa = 3.53
MM1 pKa = 7.94RR2 pKa = 11.84SQHH5 pKa = 5.94ILASSRR11 pKa = 11.84PRR13 pKa = 11.84LLAPSPPRR21 pKa = 11.84ALASSPPRR29 pKa = 11.84LLASSPPRR37 pKa = 11.84LLASSPPRR45 pKa = 11.84LLASSPPRR53 pKa = 11.84LLASSPPRR61 pKa = 11.84LLASSPPRR69 pKa = 11.84LLASSPPRR77 pKa = 11.84LL78 pKa = 3.53
Molecular weight: 8.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1557175 |
25 |
6103 |
363.2 |
39.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.698 ± 0.053 | 0.886 ± 0.013 |
4.917 ± 0.037 | 5.814 ± 0.035 |
3.328 ± 0.024 | 7.552 ± 0.044 |
2.361 ± 0.019 | 4.96 ± 0.032 |
1.94 ± 0.026 | 12.017 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.13 ± 0.016 | 2.794 ± 0.037 |
5.846 ± 0.038 | 4.537 ± 0.027 |
7.246 ± 0.04 | 5.223 ± 0.029 |
5.571 ± 0.045 | 7.051 ± 0.03 |
1.496 ± 0.017 | 2.632 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
