
Pelagibacter phage HTVC105P
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Pelagivirus; unclassified Pelagivirus
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
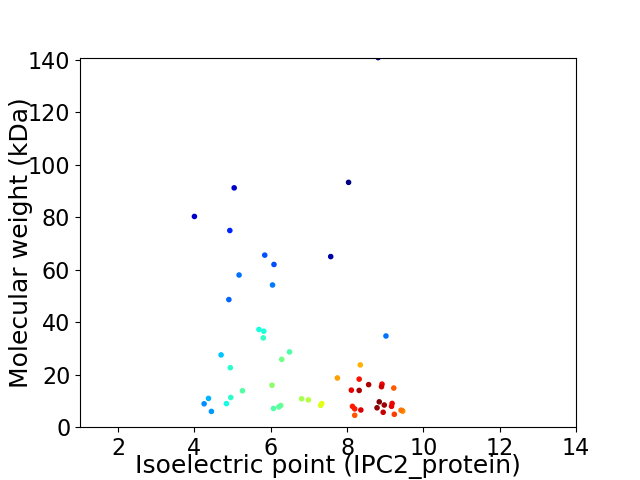
Virtual 2D-PAGE plot for 56 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y1NTB4|A0A4Y1NTB4_9CAUD Internal virion protein D OS=Pelagibacter phage HTVC105P OX=2283018 GN=P105_gp40 PE=4 SV=1
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 7.53TGNNSTTSYY17 pKa = 10.74AIPFSYY23 pKa = 10.38RR24 pKa = 11.84ATGDD28 pKa = 3.38LTVTLAGVATTAYY41 pKa = 8.61TLNAAGTTLTFNSAPAQDD59 pKa = 3.55VAIEE63 pKa = 4.07IRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.03TSQTTRR73 pKa = 11.84LTDD76 pKa = 3.51YY77 pKa = 11.47ADD79 pKa = 3.63GSVLTEE85 pKa = 4.11NDD87 pKa = 3.84LDD89 pKa = 4.08TDD91 pKa = 4.05STQAFYY97 pKa = 10.18MGQEE101 pKa = 4.74AIDD104 pKa = 4.3DD105 pKa = 4.41ANDD108 pKa = 3.41VIKK111 pKa = 10.53PSNTNFQWDD120 pKa = 3.85ANNKK124 pKa = 9.87RR125 pKa = 11.84IINVANPTSNQDD137 pKa = 3.03VATKK141 pKa = 10.18HH142 pKa = 5.5YY143 pKa = 11.03LEE145 pKa = 4.5NTWLSASDD153 pKa = 3.23KK154 pKa = 10.53TAITTVNSNIANINTVNSNITNINSAVSNATNINTVATNISSVNTVAADD203 pKa = 3.29ITKK206 pKa = 10.39VIAVANDD213 pKa = 3.2LAEE216 pKa = 4.23AVSEE220 pKa = 4.31IEE222 pKa = 4.66TVADD226 pKa = 4.16DD227 pKa = 4.47LNEE230 pKa = 4.06TSSEE234 pKa = 4.1IEE236 pKa = 3.98VVGANITNVNTVGGISANVTTVAGVSANVTTVAGISANVTTVAGISGDD284 pKa = 3.56VTAVANISSDD294 pKa = 3.32VAAVEE299 pKa = 4.2NIKK302 pKa = 10.12TNVTTVAGIASDD314 pKa = 3.68VTAVANISSDD324 pKa = 3.32VAAVEE329 pKa = 4.42NIAANVTTVAGNNANITTVAGANSNITTVAGAITNVNNVGGAITNVNNVGGSIANVNTVATNLSGVNSFAEE400 pKa = 4.56RR401 pKa = 11.84YY402 pKa = 8.58RR403 pKa = 11.84VSSSAPTSSNDD414 pKa = 3.48IGDD417 pKa = 4.59LYY419 pKa = 11.21FDD421 pKa = 3.67STANEE426 pKa = 3.89LKK428 pKa = 10.52VYY430 pKa = 10.44KK431 pKa = 10.46SSGWAAAGSTVNGTSARR448 pKa = 11.84FNYY451 pKa = 8.0TATANQTTFTGADD464 pKa = 3.17TAGNTLAYY472 pKa = 9.9DD473 pKa = 3.67AGYY476 pKa = 11.18ADD478 pKa = 4.22VYY480 pKa = 11.21LNGVRR485 pKa = 11.84LSGADD490 pKa = 3.1ITITSGTSVVLATGAAVGDD509 pKa = 3.92ILDD512 pKa = 3.71VVAYY516 pKa = 7.7GTFNVASVNAGNIDD530 pKa = 3.64AGTLNSARR538 pKa = 11.84LPTVPVSKK546 pKa = 10.85GGTNLTALGTAGQVIKK562 pKa = 11.12VNSGANALEE571 pKa = 4.13YY572 pKa = 11.19VNGSITINGSAVSLGGSVTVGEE594 pKa = 4.65TKK596 pKa = 9.07PTATGCTPSTITNSATNVVIAGTNFTSIPQVWAINTATGIWYY638 pKa = 8.88LANSVTYY645 pKa = 9.82TSATSITANFTLTVDD660 pKa = 3.32AQYY663 pKa = 10.98KK664 pKa = 9.83IRR666 pKa = 11.84VEE668 pKa = 4.11NPDD671 pKa = 3.18GNATLSTNNILTVSDD686 pKa = 3.85EE687 pKa = 4.54PTWSTSSGSLGSVAGNFSGTVATLSASSDD716 pKa = 3.49SAITYY721 pKa = 10.31SEE723 pKa = 4.18TTNVLTNASQANCALNSSTGAITTSDD749 pKa = 3.62FGGSSTTATTYY760 pKa = 11.13NFTIRR765 pKa = 11.84ATDD768 pKa = 3.74AEE770 pKa = 4.56SQTTDD775 pKa = 2.6RR776 pKa = 11.84TFSLTSSFGATGGGQFNN793 pKa = 4.04
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 7.53TGNNSTTSYY17 pKa = 10.74AIPFSYY23 pKa = 10.38RR24 pKa = 11.84ATGDD28 pKa = 3.38LTVTLAGVATTAYY41 pKa = 8.61TLNAAGTTLTFNSAPAQDD59 pKa = 3.55VAIEE63 pKa = 4.07IRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.03TSQTTRR73 pKa = 11.84LTDD76 pKa = 3.51YY77 pKa = 11.47ADD79 pKa = 3.63GSVLTEE85 pKa = 4.11NDD87 pKa = 3.84LDD89 pKa = 4.08TDD91 pKa = 4.05STQAFYY97 pKa = 10.18MGQEE101 pKa = 4.74AIDD104 pKa = 4.3DD105 pKa = 4.41ANDD108 pKa = 3.41VIKK111 pKa = 10.53PSNTNFQWDD120 pKa = 3.85ANNKK124 pKa = 9.87RR125 pKa = 11.84IINVANPTSNQDD137 pKa = 3.03VATKK141 pKa = 10.18HH142 pKa = 5.5YY143 pKa = 11.03LEE145 pKa = 4.5NTWLSASDD153 pKa = 3.23KK154 pKa = 10.53TAITTVNSNIANINTVNSNITNINSAVSNATNINTVATNISSVNTVAADD203 pKa = 3.29ITKK206 pKa = 10.39VIAVANDD213 pKa = 3.2LAEE216 pKa = 4.23AVSEE220 pKa = 4.31IEE222 pKa = 4.66TVADD226 pKa = 4.16DD227 pKa = 4.47LNEE230 pKa = 4.06TSSEE234 pKa = 4.1IEE236 pKa = 3.98VVGANITNVNTVGGISANVTTVAGVSANVTTVAGISANVTTVAGISGDD284 pKa = 3.56VTAVANISSDD294 pKa = 3.32VAAVEE299 pKa = 4.2NIKK302 pKa = 10.12TNVTTVAGIASDD314 pKa = 3.68VTAVANISSDD324 pKa = 3.32VAAVEE329 pKa = 4.42NIAANVTTVAGNNANITTVAGANSNITTVAGAITNVNNVGGAITNVNNVGGSIANVNTVATNLSGVNSFAEE400 pKa = 4.56RR401 pKa = 11.84YY402 pKa = 8.58RR403 pKa = 11.84VSSSAPTSSNDD414 pKa = 3.48IGDD417 pKa = 4.59LYY419 pKa = 11.21FDD421 pKa = 3.67STANEE426 pKa = 3.89LKK428 pKa = 10.52VYY430 pKa = 10.44KK431 pKa = 10.46SSGWAAAGSTVNGTSARR448 pKa = 11.84FNYY451 pKa = 8.0TATANQTTFTGADD464 pKa = 3.17TAGNTLAYY472 pKa = 9.9DD473 pKa = 3.67AGYY476 pKa = 11.18ADD478 pKa = 4.22VYY480 pKa = 11.21LNGVRR485 pKa = 11.84LSGADD490 pKa = 3.1ITITSGTSVVLATGAAVGDD509 pKa = 3.92ILDD512 pKa = 3.71VVAYY516 pKa = 7.7GTFNVASVNAGNIDD530 pKa = 3.64AGTLNSARR538 pKa = 11.84LPTVPVSKK546 pKa = 10.85GGTNLTALGTAGQVIKK562 pKa = 11.12VNSGANALEE571 pKa = 4.13YY572 pKa = 11.19VNGSITINGSAVSLGGSVTVGEE594 pKa = 4.65TKK596 pKa = 9.07PTATGCTPSTITNSATNVVIAGTNFTSIPQVWAINTATGIWYY638 pKa = 8.88LANSVTYY645 pKa = 9.82TSATSITANFTLTVDD660 pKa = 3.32AQYY663 pKa = 10.98KK664 pKa = 9.83IRR666 pKa = 11.84VEE668 pKa = 4.11NPDD671 pKa = 3.18GNATLSTNNILTVSDD686 pKa = 3.85EE687 pKa = 4.54PTWSTSSGSLGSVAGNFSGTVATLSASSDD716 pKa = 3.49SAITYY721 pKa = 10.31SEE723 pKa = 4.18TTNVLTNASQANCALNSSTGAITTSDD749 pKa = 3.62FGGSSTTATTYY760 pKa = 11.13NFTIRR765 pKa = 11.84ATDD768 pKa = 3.74AEE770 pKa = 4.56SQTTDD775 pKa = 2.6RR776 pKa = 11.84TFSLTSSFGATGGGQFNN793 pKa = 4.04
Molecular weight: 80.31 kDa
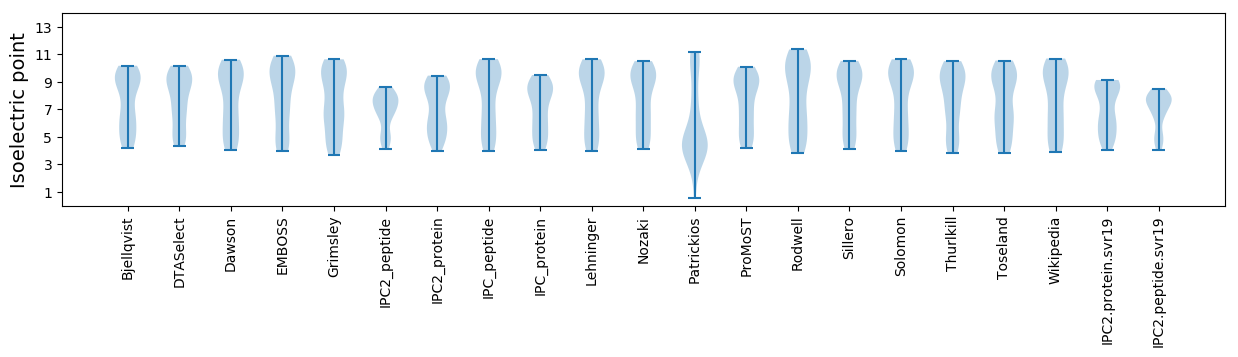
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y1NUL1|A0A4Y1NUL1_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC105P OX=2283018 GN=P105_gp37 PE=4 SV=1
MM1 pKa = 7.84TYY3 pKa = 10.24TDD5 pKa = 4.22LFKK8 pKa = 11.29DD9 pKa = 3.12VDD11 pKa = 3.64KK12 pKa = 10.8PRR14 pKa = 11.84KK15 pKa = 8.25KK16 pKa = 9.96RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.48RR21 pKa = 11.84KK22 pKa = 9.06EE23 pKa = 3.57KK24 pKa = 9.49QTVLWTVYY32 pKa = 8.84HH33 pKa = 6.8TILAVEE39 pKa = 4.29LLIIIIIEE47 pKa = 4.68GIEE50 pKa = 3.91LLRR53 pKa = 5.58
MM1 pKa = 7.84TYY3 pKa = 10.24TDD5 pKa = 4.22LFKK8 pKa = 11.29DD9 pKa = 3.12VDD11 pKa = 3.64KK12 pKa = 10.8PRR14 pKa = 11.84KK15 pKa = 8.25KK16 pKa = 9.96RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.48RR21 pKa = 11.84KK22 pKa = 9.06EE23 pKa = 3.57KK24 pKa = 9.49QTVLWTVYY32 pKa = 8.84HH33 pKa = 6.8TILAVEE39 pKa = 4.29LLIIIIIEE47 pKa = 4.68GIEE50 pKa = 3.91LLRR53 pKa = 5.58
Molecular weight: 6.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13116 |
39 |
1250 |
234.2 |
26.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.548 ± 0.493 | 0.915 ± 0.164 |
5.756 ± 0.202 | 6.862 ± 0.552 |
3.644 ± 0.211 | 6.435 ± 0.381 |
1.799 ± 0.217 | 6.496 ± 0.235 |
8.783 ± 0.638 | 7.96 ± 0.416 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.463 ± 0.23 | 5.833 ± 0.478 |
2.867 ± 0.215 | 3.614 ± 0.256 |
3.835 ± 0.249 | 6.862 ± 0.377 |
7.472 ± 0.713 | 5.932 ± 0.31 |
1.296 ± 0.118 | 3.629 ± 0.18 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
