
Roseburia sp. 831b
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Roseburia; unclassified Roseburia
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
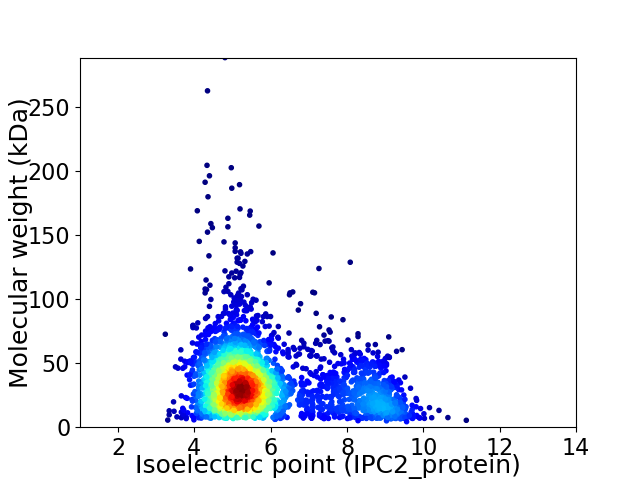
Virtual 2D-PAGE plot for 2925 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q9J6M3|A0A1Q9J6M3_9FIRM Uncharacterized protein OS=Roseburia sp. 831b OX=1261635 GN=BIV16_04395 PE=4 SV=1
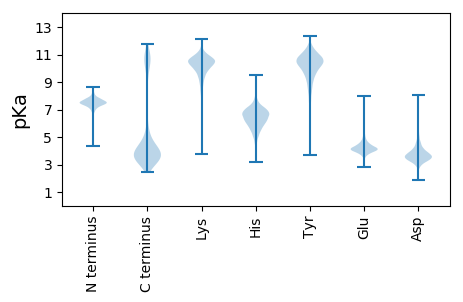
MM1 pKa = 7.43KK2 pKa = 10.33KK3 pKa = 10.34KK4 pKa = 10.01IVSVLLCVSMVATMAAGCGSSGDD27 pKa = 3.93DD28 pKa = 3.38TTTTDD33 pKa = 3.34SNSASGTTEE42 pKa = 3.71SADD45 pKa = 3.74ASGSDD50 pKa = 3.4AATGGLAYY58 pKa = 9.75TGDD61 pKa = 4.11LEE63 pKa = 4.46IMHH66 pKa = 7.06FSTEE70 pKa = 4.18EE71 pKa = 3.74EE72 pKa = 4.27SQGNGGSDD80 pKa = 3.47GMRR83 pKa = 11.84TVLQQWADD91 pKa = 3.36NNTGINLTQTVLANADD107 pKa = 3.94YY108 pKa = 8.21KK109 pKa = 10.45TQIQTLANADD119 pKa = 4.08DD120 pKa = 4.74LPDD123 pKa = 3.31VFLIQGMNVKK133 pKa = 9.21QWAEE137 pKa = 3.54QGLILDD143 pKa = 3.8LTDD146 pKa = 4.26YY147 pKa = 10.6IKK149 pKa = 10.78SSPYY153 pKa = 9.58SADD156 pKa = 3.24YY157 pKa = 10.66NEE159 pKa = 5.62SYY161 pKa = 8.02FTPFKK166 pKa = 11.1DD167 pKa = 3.31EE168 pKa = 4.41AGNIYY173 pKa = 10.08GFPALTGGTCTVVVYY188 pKa = 10.29DD189 pKa = 3.98SKK191 pKa = 11.49AWADD195 pKa = 3.26AGYY198 pKa = 8.42DD199 pKa = 3.64TFPSTWEE206 pKa = 4.06DD207 pKa = 3.38VEE209 pKa = 4.29AAIPKK214 pKa = 9.33LQEE217 pKa = 3.63AGYY220 pKa = 10.25QEE222 pKa = 5.63AIAFGNQGQWNMNSDD237 pKa = 4.33FLSTLGDD244 pKa = 3.59RR245 pKa = 11.84YY246 pKa = 8.88TGKK249 pKa = 10.47DD250 pKa = 2.97WFQSMIDD257 pKa = 3.41KK258 pKa = 10.65KK259 pKa = 11.08GAAFTDD265 pKa = 3.97DD266 pKa = 3.89EE267 pKa = 4.73FVAALTEE274 pKa = 4.2TQRR277 pKa = 11.84LFKK280 pKa = 10.63DD281 pKa = 3.09TDD283 pKa = 3.25IFNKK287 pKa = 10.24DD288 pKa = 3.04FNAITNEE295 pKa = 3.97DD296 pKa = 3.12ARR298 pKa = 11.84EE299 pKa = 3.89YY300 pKa = 11.28YY301 pKa = 9.96IGQDD305 pKa = 2.77AAAFIGGNWDD315 pKa = 2.82VSYY318 pKa = 10.78IQASLEE324 pKa = 4.08GTDD327 pKa = 5.66LYY329 pKa = 10.99DD330 pKa = 3.36TTKK333 pKa = 10.5FAVLPQPKK341 pKa = 9.28DD342 pKa = 3.28AKK344 pKa = 10.89YY345 pKa = 10.81DD346 pKa = 3.74ADD348 pKa = 3.82SQNIGLGYY356 pKa = 10.66AVAINSKK363 pKa = 10.42LADD366 pKa = 4.69DD367 pKa = 4.69PDD369 pKa = 4.17KK370 pKa = 11.19LAAAIDD376 pKa = 3.62LAEE379 pKa = 4.78YY380 pKa = 8.92ITGPEE385 pKa = 4.02FANFVCDD392 pKa = 4.22KK393 pKa = 9.99YY394 pKa = 11.74ALGGLTKK401 pKa = 10.72VNDD404 pKa = 3.26VDD406 pKa = 5.22LSKK409 pKa = 10.81FDD411 pKa = 4.32QFTQDD416 pKa = 3.53FYY418 pKa = 11.63NYY420 pKa = 9.96SYY422 pKa = 11.84VDD424 pKa = 3.43TTPCEE429 pKa = 3.97IYY431 pKa = 10.54DD432 pKa = 4.04SYY434 pKa = 11.5LSSDD438 pKa = 2.9VWSVVNTDD446 pKa = 3.43LQSMVNGDD454 pKa = 3.57MTPEE458 pKa = 3.85EE459 pKa = 4.27VAKK462 pKa = 10.4DD463 pKa = 3.41AQEE466 pKa = 4.4AYY468 pKa = 8.46EE469 pKa = 4.14ANYY472 pKa = 10.55
MM1 pKa = 7.43KK2 pKa = 10.33KK3 pKa = 10.34KK4 pKa = 10.01IVSVLLCVSMVATMAAGCGSSGDD27 pKa = 3.93DD28 pKa = 3.38TTTTDD33 pKa = 3.34SNSASGTTEE42 pKa = 3.71SADD45 pKa = 3.74ASGSDD50 pKa = 3.4AATGGLAYY58 pKa = 9.75TGDD61 pKa = 4.11LEE63 pKa = 4.46IMHH66 pKa = 7.06FSTEE70 pKa = 4.18EE71 pKa = 3.74EE72 pKa = 4.27SQGNGGSDD80 pKa = 3.47GMRR83 pKa = 11.84TVLQQWADD91 pKa = 3.36NNTGINLTQTVLANADD107 pKa = 3.94YY108 pKa = 8.21KK109 pKa = 10.45TQIQTLANADD119 pKa = 4.08DD120 pKa = 4.74LPDD123 pKa = 3.31VFLIQGMNVKK133 pKa = 9.21QWAEE137 pKa = 3.54QGLILDD143 pKa = 3.8LTDD146 pKa = 4.26YY147 pKa = 10.6IKK149 pKa = 10.78SSPYY153 pKa = 9.58SADD156 pKa = 3.24YY157 pKa = 10.66NEE159 pKa = 5.62SYY161 pKa = 8.02FTPFKK166 pKa = 11.1DD167 pKa = 3.31EE168 pKa = 4.41AGNIYY173 pKa = 10.08GFPALTGGTCTVVVYY188 pKa = 10.29DD189 pKa = 3.98SKK191 pKa = 11.49AWADD195 pKa = 3.26AGYY198 pKa = 8.42DD199 pKa = 3.64TFPSTWEE206 pKa = 4.06DD207 pKa = 3.38VEE209 pKa = 4.29AAIPKK214 pKa = 9.33LQEE217 pKa = 3.63AGYY220 pKa = 10.25QEE222 pKa = 5.63AIAFGNQGQWNMNSDD237 pKa = 4.33FLSTLGDD244 pKa = 3.59RR245 pKa = 11.84YY246 pKa = 8.88TGKK249 pKa = 10.47DD250 pKa = 2.97WFQSMIDD257 pKa = 3.41KK258 pKa = 10.65KK259 pKa = 11.08GAAFTDD265 pKa = 3.97DD266 pKa = 3.89EE267 pKa = 4.73FVAALTEE274 pKa = 4.2TQRR277 pKa = 11.84LFKK280 pKa = 10.63DD281 pKa = 3.09TDD283 pKa = 3.25IFNKK287 pKa = 10.24DD288 pKa = 3.04FNAITNEE295 pKa = 3.97DD296 pKa = 3.12ARR298 pKa = 11.84EE299 pKa = 3.89YY300 pKa = 11.28YY301 pKa = 9.96IGQDD305 pKa = 2.77AAAFIGGNWDD315 pKa = 2.82VSYY318 pKa = 10.78IQASLEE324 pKa = 4.08GTDD327 pKa = 5.66LYY329 pKa = 10.99DD330 pKa = 3.36TTKK333 pKa = 10.5FAVLPQPKK341 pKa = 9.28DD342 pKa = 3.28AKK344 pKa = 10.89YY345 pKa = 10.81DD346 pKa = 3.74ADD348 pKa = 3.82SQNIGLGYY356 pKa = 10.66AVAINSKK363 pKa = 10.42LADD366 pKa = 4.69DD367 pKa = 4.69PDD369 pKa = 4.17KK370 pKa = 11.19LAAAIDD376 pKa = 3.62LAEE379 pKa = 4.78YY380 pKa = 8.92ITGPEE385 pKa = 4.02FANFVCDD392 pKa = 4.22KK393 pKa = 9.99YY394 pKa = 11.74ALGGLTKK401 pKa = 10.72VNDD404 pKa = 3.26VDD406 pKa = 5.22LSKK409 pKa = 10.81FDD411 pKa = 4.32QFTQDD416 pKa = 3.53FYY418 pKa = 11.63NYY420 pKa = 9.96SYY422 pKa = 11.84VDD424 pKa = 3.43TTPCEE429 pKa = 3.97IYY431 pKa = 10.54DD432 pKa = 4.04SYY434 pKa = 11.5LSSDD438 pKa = 2.9VWSVVNTDD446 pKa = 3.43LQSMVNGDD454 pKa = 3.57MTPEE458 pKa = 3.85EE459 pKa = 4.27VAKK462 pKa = 10.4DD463 pKa = 3.41AQEE466 pKa = 4.4AYY468 pKa = 8.46EE469 pKa = 4.14ANYY472 pKa = 10.55
Molecular weight: 51.46 kDa
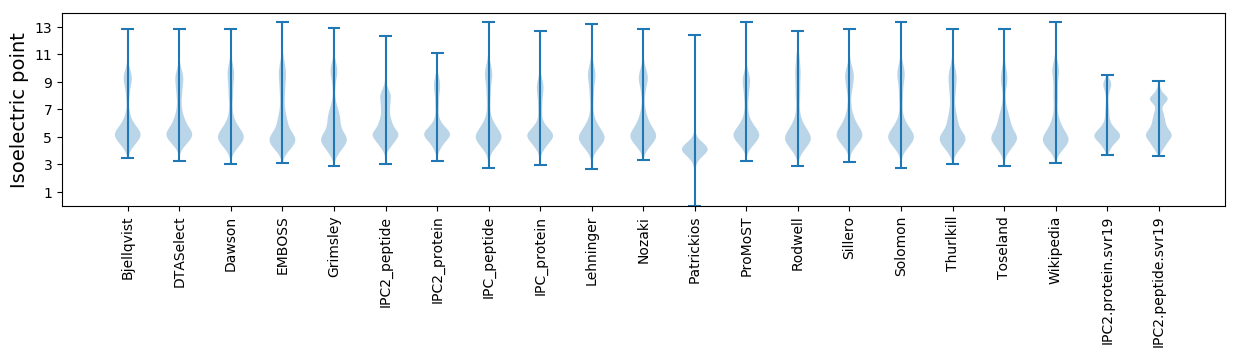
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q9J3C3|A0A1Q9J3C3_9FIRM 23S rRNA (Guanosine(2251)-2'-O)-methyltransferase RlmB OS=Roseburia sp. 831b OX=1261635 GN=BIV16_07035 PE=4 SV=1
MM1 pKa = 7.64HH2 pKa = 7.44KK3 pKa = 10.88GKK5 pKa = 8.15MTFQPKK11 pKa = 9.0NRR13 pKa = 11.84QRR15 pKa = 11.84SKK17 pKa = 8.76VHH19 pKa = 5.91GFRR22 pKa = 11.84SRR24 pKa = 11.84MSTAGGRR31 pKa = 11.84KK32 pKa = 8.62VLAARR37 pKa = 11.84RR38 pKa = 11.84LKK40 pKa = 10.61GRR42 pKa = 11.84KK43 pKa = 8.87KK44 pKa = 10.59LSAA47 pKa = 3.95
MM1 pKa = 7.64HH2 pKa = 7.44KK3 pKa = 10.88GKK5 pKa = 8.15MTFQPKK11 pKa = 9.0NRR13 pKa = 11.84QRR15 pKa = 11.84SKK17 pKa = 8.76VHH19 pKa = 5.91GFRR22 pKa = 11.84SRR24 pKa = 11.84MSTAGGRR31 pKa = 11.84KK32 pKa = 8.62VLAARR37 pKa = 11.84RR38 pKa = 11.84LKK40 pKa = 10.61GRR42 pKa = 11.84KK43 pKa = 8.87KK44 pKa = 10.59LSAA47 pKa = 3.95
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
949512 |
37 |
2533 |
324.6 |
36.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.276 ± 0.043 | 1.459 ± 0.018 |
5.781 ± 0.041 | 8.057 ± 0.055 |
4.161 ± 0.033 | 6.668 ± 0.041 |
1.709 ± 0.019 | 7.308 ± 0.047 |
7.351 ± 0.04 | 8.731 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.102 ± 0.023 | 4.597 ± 0.031 |
2.989 ± 0.026 | 3.469 ± 0.028 |
3.86 ± 0.032 | 5.773 ± 0.042 |
5.569 ± 0.044 | 7.004 ± 0.036 |
0.87 ± 0.019 | 4.266 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
