
Gila monster-associated gemycircularvirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; unclassified Gemycircularvirus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
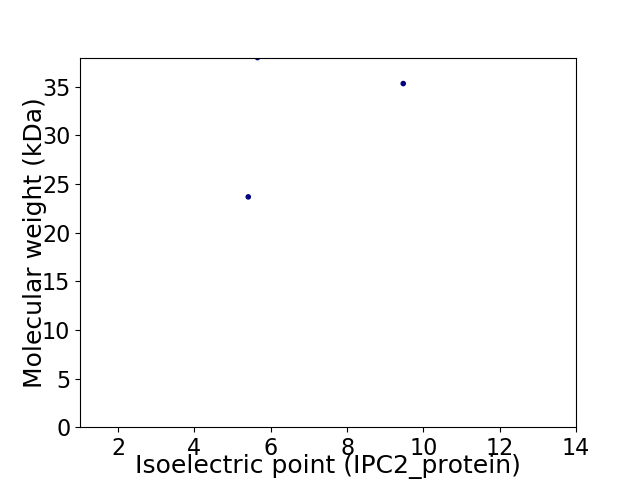
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z4N3Z3|A0A2Z4N3Z3_9VIRU Capsid protein OS=Gila monster-associated gemycircularvirus OX=2249931 PE=4 SV=1
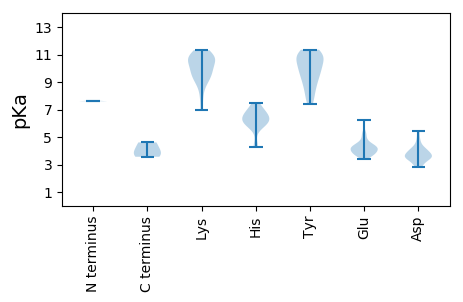
MM1 pKa = 7.6PRR3 pKa = 11.84FRR5 pKa = 11.84CRR7 pKa = 11.84ARR9 pKa = 11.84NFIITFPQVAEE20 pKa = 4.09NVQPRR25 pKa = 11.84FDD27 pKa = 4.86DD28 pKa = 3.72GTPFLEE34 pKa = 4.76RR35 pKa = 11.84VTSDD39 pKa = 3.66FGNPLCCRR47 pKa = 11.84LGRR50 pKa = 11.84EE51 pKa = 3.7RR52 pKa = 11.84HH53 pKa = 5.76ADD55 pKa = 3.38GGVHH59 pKa = 4.27YY60 pKa = 10.06HH61 pKa = 6.53VYY63 pKa = 10.77LGFDD67 pKa = 3.95KK68 pKa = 10.69IVSINSATAFDD79 pKa = 4.16YY80 pKa = 11.19FGAHH84 pKa = 6.18GNIKK88 pKa = 9.32SVRR91 pKa = 11.84RR92 pKa = 11.84TPHH95 pKa = 6.08KK96 pKa = 10.73VFDD99 pKa = 4.1YY100 pKa = 10.67VGKK103 pKa = 10.61DD104 pKa = 2.82GDD106 pKa = 3.35IRR108 pKa = 11.84LDD110 pKa = 3.41HH111 pKa = 6.83GEE113 pKa = 4.33PPAEE117 pKa = 4.09ASSRR121 pKa = 11.84DD122 pKa = 3.64GTDD125 pKa = 2.95SNKK128 pKa = 9.08WADD131 pKa = 4.41IISCDD136 pKa = 3.55TKK138 pKa = 11.11EE139 pKa = 4.29SFLSAVRR146 pKa = 11.84EE147 pKa = 3.89QAPRR151 pKa = 11.84DD152 pKa = 3.5WLLSNQRR159 pKa = 11.84ILEE162 pKa = 4.17YY163 pKa = 11.19ANLYY167 pKa = 9.47YY168 pKa = 10.01PEE170 pKa = 5.11EE171 pKa = 4.43KK172 pKa = 9.8PQYY175 pKa = 7.69EE176 pKa = 4.24APPITVEE183 pKa = 3.78RR184 pKa = 11.84EE185 pKa = 3.74RR186 pKa = 11.84YY187 pKa = 9.12PEE189 pKa = 3.64ISEE192 pKa = 4.33WEE194 pKa = 3.98NQAAIGDD201 pKa = 4.15CRR203 pKa = 11.84PEE205 pKa = 3.64RR206 pKa = 4.61
MM1 pKa = 7.6PRR3 pKa = 11.84FRR5 pKa = 11.84CRR7 pKa = 11.84ARR9 pKa = 11.84NFIITFPQVAEE20 pKa = 4.09NVQPRR25 pKa = 11.84FDD27 pKa = 4.86DD28 pKa = 3.72GTPFLEE34 pKa = 4.76RR35 pKa = 11.84VTSDD39 pKa = 3.66FGNPLCCRR47 pKa = 11.84LGRR50 pKa = 11.84EE51 pKa = 3.7RR52 pKa = 11.84HH53 pKa = 5.76ADD55 pKa = 3.38GGVHH59 pKa = 4.27YY60 pKa = 10.06HH61 pKa = 6.53VYY63 pKa = 10.77LGFDD67 pKa = 3.95KK68 pKa = 10.69IVSINSATAFDD79 pKa = 4.16YY80 pKa = 11.19FGAHH84 pKa = 6.18GNIKK88 pKa = 9.32SVRR91 pKa = 11.84RR92 pKa = 11.84TPHH95 pKa = 6.08KK96 pKa = 10.73VFDD99 pKa = 4.1YY100 pKa = 10.67VGKK103 pKa = 10.61DD104 pKa = 2.82GDD106 pKa = 3.35IRR108 pKa = 11.84LDD110 pKa = 3.41HH111 pKa = 6.83GEE113 pKa = 4.33PPAEE117 pKa = 4.09ASSRR121 pKa = 11.84DD122 pKa = 3.64GTDD125 pKa = 2.95SNKK128 pKa = 9.08WADD131 pKa = 4.41IISCDD136 pKa = 3.55TKK138 pKa = 11.11EE139 pKa = 4.29SFLSAVRR146 pKa = 11.84EE147 pKa = 3.89QAPRR151 pKa = 11.84DD152 pKa = 3.5WLLSNQRR159 pKa = 11.84ILEE162 pKa = 4.17YY163 pKa = 11.19ANLYY167 pKa = 9.47YY168 pKa = 10.01PEE170 pKa = 5.11EE171 pKa = 4.43KK172 pKa = 9.8PQYY175 pKa = 7.69EE176 pKa = 4.24APPITVEE183 pKa = 3.78RR184 pKa = 11.84EE185 pKa = 3.74RR186 pKa = 11.84YY187 pKa = 9.12PEE189 pKa = 3.64ISEE192 pKa = 4.33WEE194 pKa = 3.98NQAAIGDD201 pKa = 4.15CRR203 pKa = 11.84PEE205 pKa = 3.64RR206 pKa = 4.61
Molecular weight: 23.69 kDa
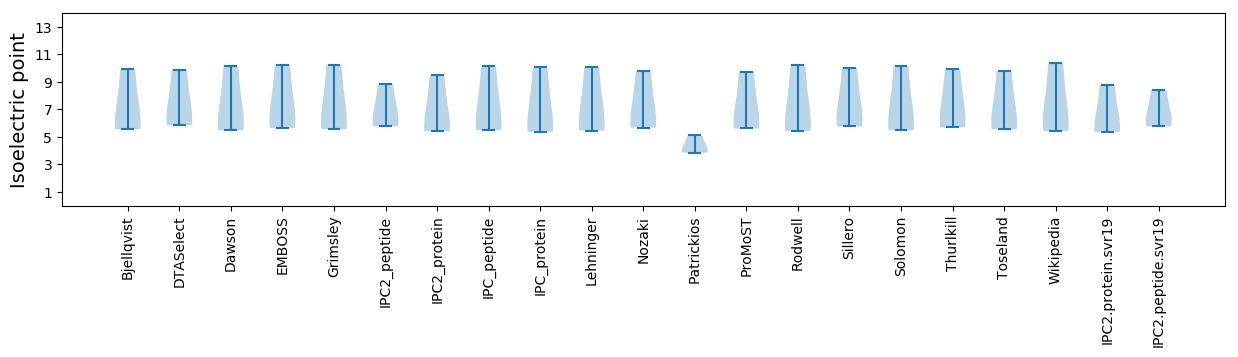
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4N3Z3|A0A2Z4N3Z3_9VIRU Capsid protein OS=Gila monster-associated gemycircularvirus OX=2249931 PE=4 SV=1
MM1 pKa = 7.64AFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84SFRR9 pKa = 11.84RR10 pKa = 11.84TGRR13 pKa = 11.84SYY15 pKa = 10.99RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84GNARR22 pKa = 11.84TSRR25 pKa = 11.84GRR27 pKa = 11.84TSGRR31 pKa = 11.84GGYY34 pKa = 7.81NRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84YY39 pKa = 7.41TGRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84KK45 pKa = 6.95MTSRR49 pKa = 11.84RR50 pKa = 11.84VRR52 pKa = 11.84DD53 pKa = 3.0IASRR57 pKa = 11.84KK58 pKa = 10.0KK59 pKa = 9.71YY60 pKa = 9.5DD61 pKa = 3.78TIFGATSNDD70 pKa = 3.49VQSTGQANIFAGTNYY85 pKa = 10.05FMWCPTWRR93 pKa = 11.84QRR95 pKa = 11.84RR96 pKa = 11.84TASTDD101 pKa = 2.79EE102 pKa = 4.09HH103 pKa = 7.51VRR105 pKa = 11.84ANQEE109 pKa = 3.49VFMRR113 pKa = 11.84GIKK116 pKa = 9.9DD117 pKa = 3.71RR118 pKa = 11.84IMVSATFSLIHH129 pKa = 6.51RR130 pKa = 11.84RR131 pKa = 11.84VCFWSHH137 pKa = 6.1RR138 pKa = 11.84RR139 pKa = 11.84IDD141 pKa = 3.56DD142 pKa = 3.43ARR144 pKa = 11.84PFVFDD149 pKa = 3.94NPDD152 pKa = 3.46VIDD155 pKa = 4.11APSYY159 pKa = 8.73QRR161 pKa = 11.84RR162 pKa = 11.84NLVALYY168 pKa = 9.61PNLDD172 pKa = 3.66EE173 pKa = 6.25EE174 pKa = 4.21IFEE177 pKa = 4.38YY178 pKa = 10.78LFKK181 pKa = 10.78GTVGFDD187 pKa = 3.3YY188 pKa = 11.33SEE190 pKa = 4.66DD191 pKa = 4.01SRR193 pKa = 11.84WDD195 pKa = 3.43TPLDD199 pKa = 3.53NKK201 pKa = 10.08RR202 pKa = 11.84VKK204 pKa = 10.23VVYY207 pKa = 10.04DD208 pKa = 3.35KK209 pKa = 11.31QYY211 pKa = 10.98TITPNYY217 pKa = 8.66AAPDD221 pKa = 3.72GAQFGKK227 pKa = 8.75TLTRR231 pKa = 11.84KK232 pKa = 9.16LWHH235 pKa = 6.6PINRR239 pKa = 11.84KK240 pKa = 8.33VLYY243 pKa = 10.66DD244 pKa = 4.14DD245 pKa = 5.44DD246 pKa = 5.45EE247 pKa = 5.3EE248 pKa = 5.35GADD251 pKa = 3.75VNGSGWSAQTPEE263 pKa = 4.28SLGNYY268 pKa = 8.41YY269 pKa = 10.94VLDD272 pKa = 3.7IFSTGQYY279 pKa = 9.92IEE281 pKa = 4.51GSNDD285 pKa = 3.24QVGTYY290 pKa = 8.5MPEE293 pKa = 3.8STTYY297 pKa = 8.23WHH299 pKa = 6.65EE300 pKa = 3.94ASS302 pKa = 3.92
MM1 pKa = 7.64AFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84SFRR9 pKa = 11.84RR10 pKa = 11.84TGRR13 pKa = 11.84SYY15 pKa = 10.99RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84GNARR22 pKa = 11.84TSRR25 pKa = 11.84GRR27 pKa = 11.84TSGRR31 pKa = 11.84GGYY34 pKa = 7.81NRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84YY39 pKa = 7.41TGRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84KK45 pKa = 6.95MTSRR49 pKa = 11.84RR50 pKa = 11.84VRR52 pKa = 11.84DD53 pKa = 3.0IASRR57 pKa = 11.84KK58 pKa = 10.0KK59 pKa = 9.71YY60 pKa = 9.5DD61 pKa = 3.78TIFGATSNDD70 pKa = 3.49VQSTGQANIFAGTNYY85 pKa = 10.05FMWCPTWRR93 pKa = 11.84QRR95 pKa = 11.84RR96 pKa = 11.84TASTDD101 pKa = 2.79EE102 pKa = 4.09HH103 pKa = 7.51VRR105 pKa = 11.84ANQEE109 pKa = 3.49VFMRR113 pKa = 11.84GIKK116 pKa = 9.9DD117 pKa = 3.71RR118 pKa = 11.84IMVSATFSLIHH129 pKa = 6.51RR130 pKa = 11.84RR131 pKa = 11.84VCFWSHH137 pKa = 6.1RR138 pKa = 11.84RR139 pKa = 11.84IDD141 pKa = 3.56DD142 pKa = 3.43ARR144 pKa = 11.84PFVFDD149 pKa = 3.94NPDD152 pKa = 3.46VIDD155 pKa = 4.11APSYY159 pKa = 8.73QRR161 pKa = 11.84RR162 pKa = 11.84NLVALYY168 pKa = 9.61PNLDD172 pKa = 3.66EE173 pKa = 6.25EE174 pKa = 4.21IFEE177 pKa = 4.38YY178 pKa = 10.78LFKK181 pKa = 10.78GTVGFDD187 pKa = 3.3YY188 pKa = 11.33SEE190 pKa = 4.66DD191 pKa = 4.01SRR193 pKa = 11.84WDD195 pKa = 3.43TPLDD199 pKa = 3.53NKK201 pKa = 10.08RR202 pKa = 11.84VKK204 pKa = 10.23VVYY207 pKa = 10.04DD208 pKa = 3.35KK209 pKa = 11.31QYY211 pKa = 10.98TITPNYY217 pKa = 8.66AAPDD221 pKa = 3.72GAQFGKK227 pKa = 8.75TLTRR231 pKa = 11.84KK232 pKa = 9.16LWHH235 pKa = 6.6PINRR239 pKa = 11.84KK240 pKa = 8.33VLYY243 pKa = 10.66DD244 pKa = 4.14DD245 pKa = 5.44DD246 pKa = 5.45EE247 pKa = 5.3EE248 pKa = 5.35GADD251 pKa = 3.75VNGSGWSAQTPEE263 pKa = 4.28SLGNYY268 pKa = 8.41YY269 pKa = 10.94VLDD272 pKa = 3.7IFSTGQYY279 pKa = 9.92IEE281 pKa = 4.51GSNDD285 pKa = 3.24QVGTYY290 pKa = 8.5MPEE293 pKa = 3.8STTYY297 pKa = 8.23WHH299 pKa = 6.65EE300 pKa = 3.94ASS302 pKa = 3.92
Molecular weight: 35.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
837 |
206 |
329 |
279.0 |
32.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.332 ± 0.274 | 1.912 ± 0.552 |
7.766 ± 0.072 | 5.974 ± 0.923 |
5.376 ± 0.201 | 6.81 ± 0.215 |
2.509 ± 0.374 | 5.496 ± 0.537 |
4.182 ± 0.423 | 4.54 ± 0.4 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.434 ± 0.303 | 4.54 ± 0.189 |
5.376 ± 0.793 | 3.106 ± 0.091 |
10.036 ± 1.5 | 6.332 ± 0.416 |
5.376 ± 1.121 | 5.615 ± 0.083 |
1.912 ± 0.187 | 5.376 ± 0.321 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
