
Beihai tombus-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
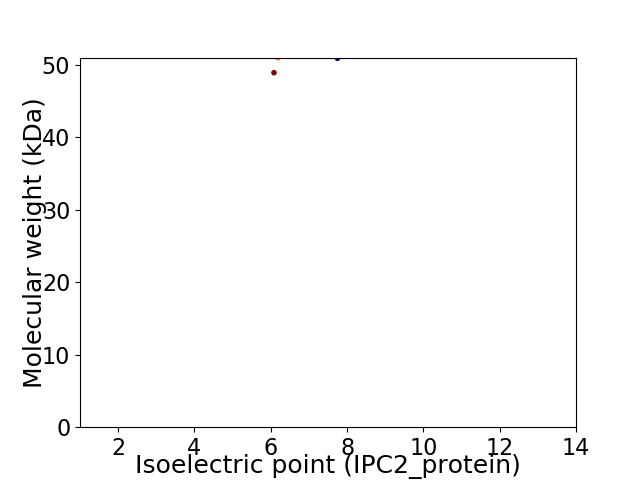
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG05|A0A1L3KG05_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 5 OX=1922726 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 9.59MYY4 pKa = 10.67ARR6 pKa = 11.84QVCRR10 pKa = 11.84GDD12 pKa = 3.3LGAITTVSFDD22 pKa = 3.21EE23 pKa = 4.57AFRR26 pKa = 11.84NFPPAKK32 pKa = 7.32RR33 pKa = 11.84QKK35 pKa = 10.0YY36 pKa = 8.38AEE38 pKa = 4.33SYY40 pKa = 10.3EE41 pKa = 4.53KK42 pKa = 10.34IGSQPDD48 pKa = 3.58MAKK51 pKa = 8.66GHH53 pKa = 6.22CMAFVKK59 pKa = 10.06YY60 pKa = 9.92EE61 pKa = 4.41RR62 pKa = 11.84LAEE65 pKa = 4.0WDD67 pKa = 3.44KK68 pKa = 11.14DD69 pKa = 3.05PRR71 pKa = 11.84MIQFRR76 pKa = 11.84NAGYY80 pKa = 9.86GAALACMLKK89 pKa = 10.24PIEE92 pKa = 3.93HH93 pKa = 7.25RR94 pKa = 11.84IYY96 pKa = 10.97KK97 pKa = 10.36LRR99 pKa = 11.84DD100 pKa = 3.56SKK102 pKa = 11.61SGLPLVAKK110 pKa = 10.2CSNAHH115 pKa = 5.42EE116 pKa = 4.16RR117 pKa = 11.84AEE119 pKa = 3.83ILRR122 pKa = 11.84EE123 pKa = 3.36KK124 pKa = 9.84WNRR127 pKa = 11.84FEE129 pKa = 4.87NPVCISYY136 pKa = 10.66DD137 pKa = 3.46CSRR140 pKa = 11.84WDD142 pKa = 3.09AHH144 pKa = 6.42VKK146 pKa = 10.11VEE148 pKa = 4.03HH149 pKa = 6.92LEE151 pKa = 4.29LEE153 pKa = 4.19HH154 pKa = 6.86EE155 pKa = 4.99FYY157 pKa = 11.62SMCNPDD163 pKa = 3.67PQLPTLLQKK172 pKa = 9.11QLRR175 pKa = 11.84NSVFTANGLFYY186 pKa = 10.51QVLGGRR192 pKa = 11.84MSGDD196 pKa = 3.45MNTSSGNVALMLFFMTAATKK216 pKa = 10.02GFRR219 pKa = 11.84RR220 pKa = 11.84EE221 pKa = 4.04IYY223 pKa = 10.38DD224 pKa = 4.65DD225 pKa = 5.05GDD227 pKa = 3.78DD228 pKa = 3.6CVVIVEE234 pKa = 4.84AEE236 pKa = 4.1DD237 pKa = 3.59EE238 pKa = 4.44EE239 pKa = 5.17KK240 pKa = 10.54ISEE243 pKa = 4.47RR244 pKa = 11.84VLEE247 pKa = 4.34WFTQAGFVIRR257 pKa = 11.84VDD259 pKa = 3.22AVARR263 pKa = 11.84QFEE266 pKa = 4.81EE267 pKa = 5.01IMFCSCHH274 pKa = 5.94PVWDD278 pKa = 4.0GQIYY282 pKa = 10.87KK283 pKa = 9.1MVRR286 pKa = 11.84DD287 pKa = 3.47WRR289 pKa = 11.84KK290 pKa = 9.02VVGFGISGAKK300 pKa = 8.78WMRR303 pKa = 11.84QGEE306 pKa = 4.55KK307 pKa = 10.78AMKK310 pKa = 9.69TYY312 pKa = 10.5IRR314 pKa = 11.84TNGVCEE320 pKa = 3.77MALTRR325 pKa = 11.84GVPVLQTYY333 pKa = 7.79AQRR336 pKa = 11.84LIQFGQGGKK345 pKa = 9.85LVEE348 pKa = 4.75DD349 pKa = 3.26AMEE352 pKa = 4.11VKK354 pKa = 10.33KK355 pKa = 10.61RR356 pKa = 11.84LRR358 pKa = 11.84LAMSNDD364 pKa = 3.2QVEE367 pKa = 4.53SLDD370 pKa = 4.93QIDD373 pKa = 3.85TTPRR377 pKa = 11.84DD378 pKa = 3.62ITWEE382 pKa = 3.99SRR384 pKa = 11.84EE385 pKa = 4.01SFAIAFNINPQQQIDD400 pKa = 3.88MEE402 pKa = 4.39QTISNWTFPQIEE414 pKa = 4.31QCVLPEE420 pKa = 3.8WRR422 pKa = 11.84TSS424 pKa = 3.06
MM1 pKa = 7.43KK2 pKa = 9.59MYY4 pKa = 10.67ARR6 pKa = 11.84QVCRR10 pKa = 11.84GDD12 pKa = 3.3LGAITTVSFDD22 pKa = 3.21EE23 pKa = 4.57AFRR26 pKa = 11.84NFPPAKK32 pKa = 7.32RR33 pKa = 11.84QKK35 pKa = 10.0YY36 pKa = 8.38AEE38 pKa = 4.33SYY40 pKa = 10.3EE41 pKa = 4.53KK42 pKa = 10.34IGSQPDD48 pKa = 3.58MAKK51 pKa = 8.66GHH53 pKa = 6.22CMAFVKK59 pKa = 10.06YY60 pKa = 9.92EE61 pKa = 4.41RR62 pKa = 11.84LAEE65 pKa = 4.0WDD67 pKa = 3.44KK68 pKa = 11.14DD69 pKa = 3.05PRR71 pKa = 11.84MIQFRR76 pKa = 11.84NAGYY80 pKa = 9.86GAALACMLKK89 pKa = 10.24PIEE92 pKa = 3.93HH93 pKa = 7.25RR94 pKa = 11.84IYY96 pKa = 10.97KK97 pKa = 10.36LRR99 pKa = 11.84DD100 pKa = 3.56SKK102 pKa = 11.61SGLPLVAKK110 pKa = 10.2CSNAHH115 pKa = 5.42EE116 pKa = 4.16RR117 pKa = 11.84AEE119 pKa = 3.83ILRR122 pKa = 11.84EE123 pKa = 3.36KK124 pKa = 9.84WNRR127 pKa = 11.84FEE129 pKa = 4.87NPVCISYY136 pKa = 10.66DD137 pKa = 3.46CSRR140 pKa = 11.84WDD142 pKa = 3.09AHH144 pKa = 6.42VKK146 pKa = 10.11VEE148 pKa = 4.03HH149 pKa = 6.92LEE151 pKa = 4.29LEE153 pKa = 4.19HH154 pKa = 6.86EE155 pKa = 4.99FYY157 pKa = 11.62SMCNPDD163 pKa = 3.67PQLPTLLQKK172 pKa = 9.11QLRR175 pKa = 11.84NSVFTANGLFYY186 pKa = 10.51QVLGGRR192 pKa = 11.84MSGDD196 pKa = 3.45MNTSSGNVALMLFFMTAATKK216 pKa = 10.02GFRR219 pKa = 11.84RR220 pKa = 11.84EE221 pKa = 4.04IYY223 pKa = 10.38DD224 pKa = 4.65DD225 pKa = 5.05GDD227 pKa = 3.78DD228 pKa = 3.6CVVIVEE234 pKa = 4.84AEE236 pKa = 4.1DD237 pKa = 3.59EE238 pKa = 4.44EE239 pKa = 5.17KK240 pKa = 10.54ISEE243 pKa = 4.47RR244 pKa = 11.84VLEE247 pKa = 4.34WFTQAGFVIRR257 pKa = 11.84VDD259 pKa = 3.22AVARR263 pKa = 11.84QFEE266 pKa = 4.81EE267 pKa = 5.01IMFCSCHH274 pKa = 5.94PVWDD278 pKa = 4.0GQIYY282 pKa = 10.87KK283 pKa = 9.1MVRR286 pKa = 11.84DD287 pKa = 3.47WRR289 pKa = 11.84KK290 pKa = 9.02VVGFGISGAKK300 pKa = 8.78WMRR303 pKa = 11.84QGEE306 pKa = 4.55KK307 pKa = 10.78AMKK310 pKa = 9.69TYY312 pKa = 10.5IRR314 pKa = 11.84TNGVCEE320 pKa = 3.77MALTRR325 pKa = 11.84GVPVLQTYY333 pKa = 7.79AQRR336 pKa = 11.84LIQFGQGGKK345 pKa = 9.85LVEE348 pKa = 4.75DD349 pKa = 3.26AMEE352 pKa = 4.11VKK354 pKa = 10.33KK355 pKa = 10.61RR356 pKa = 11.84LRR358 pKa = 11.84LAMSNDD364 pKa = 3.2QVEE367 pKa = 4.53SLDD370 pKa = 4.93QIDD373 pKa = 3.85TTPRR377 pKa = 11.84DD378 pKa = 3.62ITWEE382 pKa = 3.99SRR384 pKa = 11.84EE385 pKa = 4.01SFAIAFNINPQQQIDD400 pKa = 3.88MEE402 pKa = 4.39QTISNWTFPQIEE414 pKa = 4.31QCVLPEE420 pKa = 3.8WRR422 pKa = 11.84TSS424 pKa = 3.06
Molecular weight: 48.93 kDa
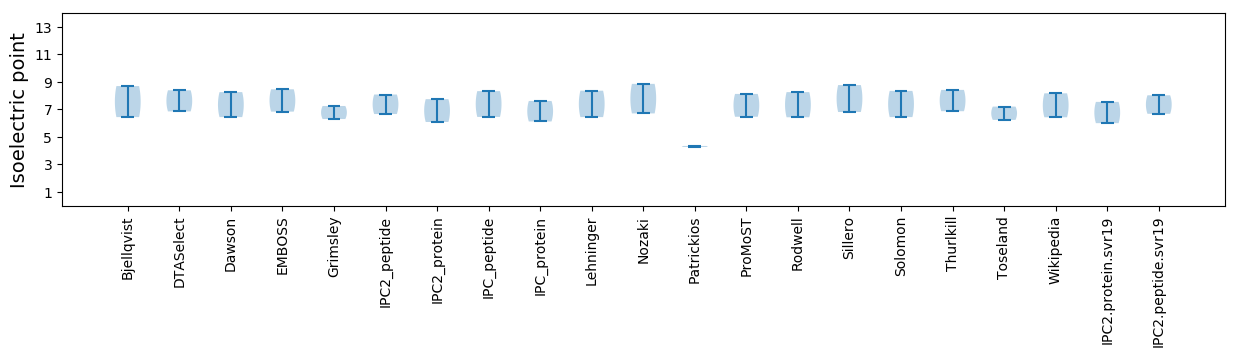
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG05|A0A1L3KG05_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 5 OX=1922726 PE=4 SV=1
MM1 pKa = 7.25LRR3 pKa = 11.84EE4 pKa = 4.44HH5 pKa = 6.79APEE8 pKa = 4.33MGTIMYY14 pKa = 10.39HH15 pKa = 6.42NGDD18 pKa = 3.44NGLNRR23 pKa = 11.84VLAVVCTTRR32 pKa = 11.84PAMDD36 pKa = 4.28LAQRR40 pKa = 11.84LRR42 pKa = 11.84EE43 pKa = 3.84IDD45 pKa = 4.27LLQRR49 pKa = 11.84WDD51 pKa = 3.72GPVYY55 pKa = 10.46IVPQRR60 pKa = 11.84IGRR63 pKa = 11.84FSAPQFIEE71 pKa = 4.12TCRR74 pKa = 11.84VAMPQCVVARR84 pKa = 11.84FYY86 pKa = 10.69PPRR89 pKa = 11.84RR90 pKa = 11.84LQQEE94 pKa = 4.15VEE96 pKa = 4.03EE97 pKa = 4.81ANQNDD102 pKa = 4.19AGEE105 pKa = 4.46EE106 pKa = 3.79PAVAQVEE113 pKa = 4.31RR114 pKa = 11.84QEE116 pKa = 4.44PDD118 pKa = 3.31DD119 pKa = 4.72NEE121 pKa = 3.82VDD123 pKa = 3.73EE124 pKa = 4.85NMEE127 pKa = 4.85LGPDD131 pKa = 3.22HH132 pKa = 6.85HH133 pKa = 8.39VPVDD137 pKa = 3.44DD138 pKa = 3.82ATLTGRR144 pKa = 11.84LATWWLRR151 pKa = 11.84ITRR154 pKa = 11.84QLRR157 pKa = 11.84QPEE160 pKa = 4.33VPADD164 pKa = 3.74VRR166 pKa = 11.84WARR169 pKa = 11.84CGVFPIINLRR179 pKa = 11.84ATGSGLMLVGSALAAANVPRR199 pKa = 11.84LTWEE203 pKa = 3.89LSYY206 pKa = 11.21NYY208 pKa = 9.72PCEE211 pKa = 4.2LTSKK215 pKa = 11.02ALATFCNATNVTLDD229 pKa = 3.25NALVHH234 pKa = 6.52HH235 pKa = 7.14IKK237 pKa = 10.25PYY239 pKa = 8.98SHH241 pKa = 7.39PAFHH245 pKa = 6.57ATYY248 pKa = 8.32LTRR251 pKa = 11.84VIKK254 pKa = 10.19FVSKK258 pKa = 10.85NIPLLANTSIGWTMRR273 pKa = 11.84KK274 pKa = 9.02CRR276 pKa = 11.84CTGPWYY282 pKa = 10.07YY283 pKa = 10.0PKK285 pKa = 10.52PKK287 pKa = 9.77WSWLGPAVFVIGCGLLYY304 pKa = 10.32IGVRR308 pKa = 11.84AIVRR312 pKa = 11.84SDD314 pKa = 3.6PEE316 pKa = 5.0ADD318 pKa = 3.05AVARR322 pKa = 11.84QAAFMRR328 pKa = 11.84PRR330 pKa = 11.84NRR332 pKa = 11.84NTLTVIEE339 pKa = 4.6GRR341 pKa = 11.84LDD343 pKa = 2.84AWMQRR348 pKa = 11.84EE349 pKa = 4.13NVPIVYY355 pKa = 10.02RR356 pKa = 11.84AGVRR360 pKa = 11.84VASAVRR366 pKa = 11.84AMDD369 pKa = 3.41VSPEE373 pKa = 4.03EE374 pKa = 4.1EE375 pKa = 4.01HH376 pKa = 7.48LLTQVGGVQQNLRR389 pKa = 11.84NADD392 pKa = 3.27VSGWIGNVSQYY403 pKa = 10.98LRR405 pKa = 11.84PFLPFLGVGLGLVYY419 pKa = 9.87FRR421 pKa = 11.84PPQSVLVAGRR431 pKa = 11.84SSLYY435 pKa = 11.29AMGQEE440 pKa = 4.97LIRR443 pKa = 11.84PGLQPTLTGGLHH455 pKa = 5.93
MM1 pKa = 7.25LRR3 pKa = 11.84EE4 pKa = 4.44HH5 pKa = 6.79APEE8 pKa = 4.33MGTIMYY14 pKa = 10.39HH15 pKa = 6.42NGDD18 pKa = 3.44NGLNRR23 pKa = 11.84VLAVVCTTRR32 pKa = 11.84PAMDD36 pKa = 4.28LAQRR40 pKa = 11.84LRR42 pKa = 11.84EE43 pKa = 3.84IDD45 pKa = 4.27LLQRR49 pKa = 11.84WDD51 pKa = 3.72GPVYY55 pKa = 10.46IVPQRR60 pKa = 11.84IGRR63 pKa = 11.84FSAPQFIEE71 pKa = 4.12TCRR74 pKa = 11.84VAMPQCVVARR84 pKa = 11.84FYY86 pKa = 10.69PPRR89 pKa = 11.84RR90 pKa = 11.84LQQEE94 pKa = 4.15VEE96 pKa = 4.03EE97 pKa = 4.81ANQNDD102 pKa = 4.19AGEE105 pKa = 4.46EE106 pKa = 3.79PAVAQVEE113 pKa = 4.31RR114 pKa = 11.84QEE116 pKa = 4.44PDD118 pKa = 3.31DD119 pKa = 4.72NEE121 pKa = 3.82VDD123 pKa = 3.73EE124 pKa = 4.85NMEE127 pKa = 4.85LGPDD131 pKa = 3.22HH132 pKa = 6.85HH133 pKa = 8.39VPVDD137 pKa = 3.44DD138 pKa = 3.82ATLTGRR144 pKa = 11.84LATWWLRR151 pKa = 11.84ITRR154 pKa = 11.84QLRR157 pKa = 11.84QPEE160 pKa = 4.33VPADD164 pKa = 3.74VRR166 pKa = 11.84WARR169 pKa = 11.84CGVFPIINLRR179 pKa = 11.84ATGSGLMLVGSALAAANVPRR199 pKa = 11.84LTWEE203 pKa = 3.89LSYY206 pKa = 11.21NYY208 pKa = 9.72PCEE211 pKa = 4.2LTSKK215 pKa = 11.02ALATFCNATNVTLDD229 pKa = 3.25NALVHH234 pKa = 6.52HH235 pKa = 7.14IKK237 pKa = 10.25PYY239 pKa = 8.98SHH241 pKa = 7.39PAFHH245 pKa = 6.57ATYY248 pKa = 8.32LTRR251 pKa = 11.84VIKK254 pKa = 10.19FVSKK258 pKa = 10.85NIPLLANTSIGWTMRR273 pKa = 11.84KK274 pKa = 9.02CRR276 pKa = 11.84CTGPWYY282 pKa = 10.07YY283 pKa = 10.0PKK285 pKa = 10.52PKK287 pKa = 9.77WSWLGPAVFVIGCGLLYY304 pKa = 10.32IGVRR308 pKa = 11.84AIVRR312 pKa = 11.84SDD314 pKa = 3.6PEE316 pKa = 5.0ADD318 pKa = 3.05AVARR322 pKa = 11.84QAAFMRR328 pKa = 11.84PRR330 pKa = 11.84NRR332 pKa = 11.84NTLTVIEE339 pKa = 4.6GRR341 pKa = 11.84LDD343 pKa = 2.84AWMQRR348 pKa = 11.84EE349 pKa = 4.13NVPIVYY355 pKa = 10.02RR356 pKa = 11.84AGVRR360 pKa = 11.84VASAVRR366 pKa = 11.84AMDD369 pKa = 3.41VSPEE373 pKa = 4.03EE374 pKa = 4.1EE375 pKa = 4.01HH376 pKa = 7.48LLTQVGGVQQNLRR389 pKa = 11.84NADD392 pKa = 3.27VSGWIGNVSQYY403 pKa = 10.98LRR405 pKa = 11.84PFLPFLGVGLGLVYY419 pKa = 9.87FRR421 pKa = 11.84PPQSVLVAGRR431 pKa = 11.84SSLYY435 pKa = 11.29AMGQEE440 pKa = 4.97LIRR443 pKa = 11.84PGLQPTLTGGLHH455 pKa = 5.93
Molecular weight: 50.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
879 |
424 |
455 |
439.5 |
49.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.419 ± 0.605 | 2.389 ± 0.306 |
4.778 ± 0.613 | 6.485 ± 0.901 |
3.754 ± 0.832 | 6.485 ± 0.409 |
1.934 ± 0.197 | 4.778 ± 0.449 |
3.527 ± 1.481 | 8.191 ± 1.266 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.527 ± 0.663 | 4.096 ± 0.387 |
5.688 ± 1.329 | 5.119 ± 0.376 |
8.077 ± 0.532 | 4.437 ± 0.522 |
4.778 ± 0.37 | 8.077 ± 1.023 |
2.389 ± 0.021 | 3.072 ± 0.004 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
