
Groundnut chlorotic fan-spot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Tospoviridae; Orthotospovirus; Groundnut chlorotic fan spot orthotospovirus
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
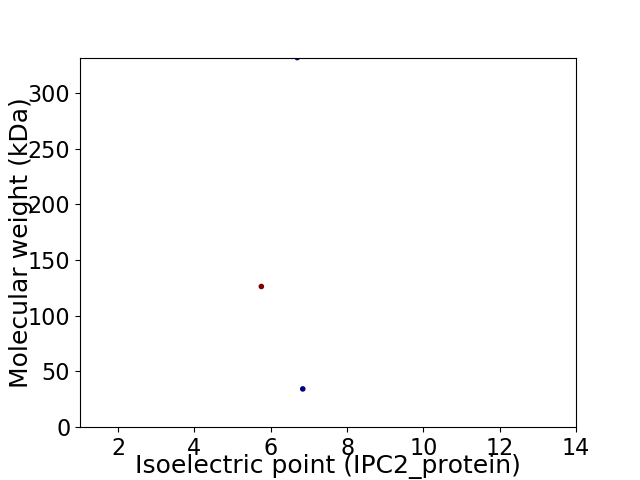
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D4D8M7|A0A0D4D8M7_9VIRU Envelopment polyprotein OS=Groundnut chlorotic fan-spot virus OX=1629551 PE=4 SV=1
MM1 pKa = 7.42SSSLPIYY8 pKa = 10.14LSGFIFYY15 pKa = 8.12VTSLLVIMILMLCLILNLRR34 pKa = 11.84QLSSSEE40 pKa = 4.03KK41 pKa = 10.05SPSLEE46 pKa = 3.96SQSGKK51 pKa = 10.31KK52 pKa = 9.76KK53 pKa = 10.23YY54 pKa = 10.05SGTYY58 pKa = 9.1RR59 pKa = 11.84SVMLSDD65 pKa = 4.28SYY67 pKa = 11.42TDD69 pKa = 3.71KK70 pKa = 11.02EE71 pKa = 4.01KK72 pKa = 10.93CRR74 pKa = 11.84YY75 pKa = 7.93TEE77 pKa = 4.13QNHH80 pKa = 5.58EE81 pKa = 4.34PPLPSNCMIVGKK93 pKa = 8.23STFNFLATMNEE104 pKa = 3.46GDD106 pKa = 5.29DD107 pKa = 4.92YY108 pKa = 10.43YY109 pKa = 10.89TCPSEE114 pKa = 5.09NVTFYY119 pKa = 11.13KK120 pKa = 10.1KK121 pKa = 10.48CKK123 pKa = 10.08NINLLSDD130 pKa = 4.38FNSEE134 pKa = 3.25ITIPVVPVMNVEE146 pKa = 4.14NKK148 pKa = 10.05RR149 pKa = 11.84LLEE152 pKa = 4.23VGSKK156 pKa = 10.43YY157 pKa = 10.5FEE159 pKa = 4.06EE160 pKa = 5.53KK161 pKa = 10.16IGSKK165 pKa = 7.26QHH167 pKa = 6.01IVLHH171 pKa = 5.18SVKK174 pKa = 10.65LSGDD178 pKa = 3.82CNISQLNSDD187 pKa = 3.37EE188 pKa = 4.35TFNVNLEE195 pKa = 4.3TTEE198 pKa = 3.95KK199 pKa = 10.85LIGVTVKK206 pKa = 10.63QMGQLDD212 pKa = 4.04SKK214 pKa = 10.31VQSFTGTFQYY224 pKa = 10.83QLSEE228 pKa = 4.26DD229 pKa = 4.01SLDD232 pKa = 3.53GFHH235 pKa = 7.21YY236 pKa = 9.39LICGDD241 pKa = 4.42KK242 pKa = 10.48FFHH245 pKa = 6.88LPNVDD250 pKa = 3.14NGLRR254 pKa = 11.84RR255 pKa = 11.84CVHH258 pKa = 7.01KK259 pKa = 11.21YY260 pKa = 9.73QDD262 pKa = 4.21NKK264 pKa = 9.93TMLYY268 pKa = 9.01TCVNFYY274 pKa = 8.39WLKK277 pKa = 8.82WAFLFFLIMMPITWIMNKK295 pKa = 8.77TKK297 pKa = 10.69GPLSLWYY304 pKa = 9.74DD305 pKa = 3.75LLSIVLYY312 pKa = 9.51PLIYY316 pKa = 10.55LLNNLWSKK324 pKa = 9.82MPYY327 pKa = 9.1RR328 pKa = 11.84CDD330 pKa = 2.98MCGYY334 pKa = 10.67LSLLSHH340 pKa = 7.47DD341 pKa = 4.91CPPKK345 pKa = 10.07CVCGRR350 pKa = 11.84SDD352 pKa = 4.59SGALHH357 pKa = 6.99NPDD360 pKa = 4.35CYY362 pKa = 10.83FNRR365 pKa = 11.84GFPKK369 pKa = 10.39KK370 pKa = 9.63PSKK373 pKa = 10.57KK374 pKa = 10.07FQFLMNTGITRR385 pKa = 11.84NVTFYY390 pKa = 10.04LTRR393 pKa = 11.84FMLSTIVISILPNTLALQASCGYY416 pKa = 10.08NCYY419 pKa = 10.82ADD421 pKa = 5.08LEE423 pKa = 4.42QSKK426 pKa = 10.0FLYY429 pKa = 9.98IGSHH433 pKa = 5.36SMPSARR439 pKa = 11.84EE440 pKa = 3.91CDD442 pKa = 3.59CKK444 pKa = 10.01ITKK447 pKa = 10.33DD448 pKa = 3.55SGSLDD453 pKa = 3.53FPFKK457 pKa = 10.85VVEE460 pKa = 4.43TYY462 pKa = 11.29KK463 pKa = 10.96DD464 pKa = 3.3ITGKK468 pKa = 8.02TQTNIANKK476 pKa = 9.8KK477 pKa = 9.5CYY479 pKa = 9.61PDD481 pKa = 5.04FDD483 pKa = 5.89DD484 pKa = 5.96DD485 pKa = 5.13CADD488 pKa = 3.77HH489 pKa = 7.39LDD491 pKa = 4.0TEE493 pKa = 4.51VLRR496 pKa = 11.84ILACAYY502 pKa = 10.27SCEE505 pKa = 3.92ALKK508 pKa = 10.2MYY510 pKa = 9.94IYY512 pKa = 10.54EE513 pKa = 4.46PLPSVHH519 pKa = 6.58EE520 pKa = 4.49SYY522 pKa = 10.68KK523 pKa = 10.81GEE525 pKa = 4.38AYY527 pKa = 10.6GKK529 pKa = 8.49MKK531 pKa = 10.47PIDD534 pKa = 4.32LYY536 pKa = 11.42SLQKK540 pKa = 10.69SRR542 pKa = 11.84TGVIDD547 pKa = 3.09NRR549 pKa = 11.84TILSRR554 pKa = 11.84QEE556 pKa = 4.0PSLSTWEE563 pKa = 4.08KK564 pKa = 9.09TNSVMSKK571 pKa = 10.62AIASLDD577 pKa = 3.61KK578 pKa = 11.07LDD580 pKa = 3.87VSSISDD586 pKa = 3.7SNTLSRR592 pKa = 11.84TTLMYY597 pKa = 10.41SSKK600 pKa = 10.69VDD602 pKa = 3.19GKK604 pKa = 9.28YY605 pKa = 10.28RR606 pKa = 11.84YY607 pKa = 9.32MIEE610 pKa = 3.75QDD612 pKa = 4.5LISDD616 pKa = 4.15TGTVFEE622 pKa = 4.85MVDD625 pKa = 3.24GTMSHH630 pKa = 6.04SKK632 pKa = 10.93KK633 pKa = 10.94LMITHH638 pKa = 6.74KK639 pKa = 10.51NVGVTYY645 pKa = 9.4TIEE648 pKa = 4.01YY649 pKa = 9.92LYY651 pKa = 8.39TTADD655 pKa = 2.85IVTTKK660 pKa = 10.25TSIYY664 pKa = 10.27SSCTGNCEE672 pKa = 4.16KK673 pKa = 10.48CWEE676 pKa = 4.39GLGSTDD682 pKa = 2.96NPSYY686 pKa = 11.09HH687 pKa = 6.68KK688 pKa = 10.62FCLEE692 pKa = 3.85PTSQWGCEE700 pKa = 3.94DD701 pKa = 3.9FMCIAMNEE709 pKa = 4.14GSTCGHH715 pKa = 7.1CDD717 pKa = 3.06NVYY720 pKa = 11.44DD721 pKa = 4.29MSNSLDD727 pKa = 3.47VYY729 pKa = 9.88RR730 pKa = 11.84VKK732 pKa = 10.43EE733 pKa = 3.88VHH735 pKa = 6.32VSSTACVNAGKK746 pKa = 10.6GYY748 pKa = 10.65NCFNHH753 pKa = 7.67DD754 pKa = 4.28DD755 pKa = 3.82RR756 pKa = 11.84KK757 pKa = 10.95AYY759 pKa = 9.75TNDD762 pKa = 3.14NYY764 pKa = 10.6QVSITSTLQNDD775 pKa = 4.66YY776 pKa = 11.0ISVGHH781 pKa = 6.74KK782 pKa = 10.06LALAKK787 pKa = 10.71DD788 pKa = 4.17GSAYY792 pKa = 9.42TGNIADD798 pKa = 4.91LMSADD803 pKa = 3.69MTFGHH808 pKa = 6.59PQLNKK813 pKa = 10.11QGEE816 pKa = 4.28LLFAEE821 pKa = 4.93EE822 pKa = 4.63YY823 pKa = 9.98IGKK826 pKa = 8.58EE827 pKa = 3.75QLSWDD832 pKa = 4.18CEE834 pKa = 4.31FIGPKK839 pKa = 9.53KK840 pKa = 10.89VYY842 pKa = 9.54IKK844 pKa = 10.75KK845 pKa = 10.26CGFYY849 pKa = 10.39TFDD852 pKa = 3.89HH853 pKa = 6.65FSGLEE858 pKa = 3.76KK859 pKa = 10.72LNEE862 pKa = 4.01GSFVITGLNKK872 pKa = 8.67ITMLKK877 pKa = 10.25DD878 pKa = 3.25YY879 pKa = 10.91SLGKK883 pKa = 10.18IKK885 pKa = 9.72TVLDD889 pKa = 3.74LPIEE893 pKa = 4.21LFVEE897 pKa = 4.4TTEE900 pKa = 4.08APKK903 pKa = 11.11VNIIDD908 pKa = 4.29HH909 pKa = 6.24FCSGCLEE916 pKa = 4.28CFKK919 pKa = 11.28GLDD922 pKa = 3.66CKK924 pKa = 11.31YY925 pKa = 10.44KK926 pKa = 10.49INSNKK931 pKa = 9.57HH932 pKa = 3.82FASRR936 pKa = 11.84ISSDD940 pKa = 3.46FCVSKK945 pKa = 10.24HH946 pKa = 5.31QYY948 pKa = 10.65LIVEE952 pKa = 4.7RR953 pKa = 11.84GSNEE957 pKa = 2.98ISMNLHH963 pKa = 6.33CGVLPEE969 pKa = 3.99NAVLKK974 pKa = 10.01MIVEE978 pKa = 4.44DD979 pKa = 4.2NEE981 pKa = 4.52NISLDD986 pKa = 3.37IPLTKK991 pKa = 9.67IEE993 pKa = 4.98IINDD997 pKa = 3.52KK998 pKa = 10.83NVEE1001 pKa = 4.13TLKK1004 pKa = 10.73DD1005 pKa = 3.33HH1006 pKa = 7.68RR1007 pKa = 11.84IQKK1010 pKa = 9.67SFQHH1014 pKa = 6.9KK1015 pKa = 10.11EE1016 pKa = 3.67DD1017 pKa = 3.85TDD1019 pKa = 3.76FHH1021 pKa = 6.46TFLEE1025 pKa = 4.86TVTYY1029 pKa = 9.07PFNWVANFTNSALDD1043 pKa = 3.9LLKK1046 pKa = 10.81LALLFGFIILGLYY1059 pKa = 9.45VLSKK1063 pKa = 10.6CVNLGKK1069 pKa = 9.94EE1070 pKa = 4.44AYY1072 pKa = 9.26SHH1074 pKa = 5.29ATIYY1078 pKa = 10.27IKK1080 pKa = 10.53KK1081 pKa = 9.83KK1082 pKa = 10.2KK1083 pKa = 9.85HH1084 pKa = 6.8DD1085 pKa = 4.91DD1086 pKa = 3.56NDD1088 pKa = 4.23SDD1090 pKa = 4.21VDD1092 pKa = 3.76DD1093 pKa = 4.63SEE1095 pKa = 4.92MYY1097 pKa = 10.71GEE1099 pKa = 4.33KK1100 pKa = 10.71NLVRR1104 pKa = 11.84RR1105 pKa = 11.84LKK1107 pKa = 11.11YY1108 pKa = 9.83IDD1110 pKa = 3.53LL1111 pKa = 4.45
MM1 pKa = 7.42SSSLPIYY8 pKa = 10.14LSGFIFYY15 pKa = 8.12VTSLLVIMILMLCLILNLRR34 pKa = 11.84QLSSSEE40 pKa = 4.03KK41 pKa = 10.05SPSLEE46 pKa = 3.96SQSGKK51 pKa = 10.31KK52 pKa = 9.76KK53 pKa = 10.23YY54 pKa = 10.05SGTYY58 pKa = 9.1RR59 pKa = 11.84SVMLSDD65 pKa = 4.28SYY67 pKa = 11.42TDD69 pKa = 3.71KK70 pKa = 11.02EE71 pKa = 4.01KK72 pKa = 10.93CRR74 pKa = 11.84YY75 pKa = 7.93TEE77 pKa = 4.13QNHH80 pKa = 5.58EE81 pKa = 4.34PPLPSNCMIVGKK93 pKa = 8.23STFNFLATMNEE104 pKa = 3.46GDD106 pKa = 5.29DD107 pKa = 4.92YY108 pKa = 10.43YY109 pKa = 10.89TCPSEE114 pKa = 5.09NVTFYY119 pKa = 11.13KK120 pKa = 10.1KK121 pKa = 10.48CKK123 pKa = 10.08NINLLSDD130 pKa = 4.38FNSEE134 pKa = 3.25ITIPVVPVMNVEE146 pKa = 4.14NKK148 pKa = 10.05RR149 pKa = 11.84LLEE152 pKa = 4.23VGSKK156 pKa = 10.43YY157 pKa = 10.5FEE159 pKa = 4.06EE160 pKa = 5.53KK161 pKa = 10.16IGSKK165 pKa = 7.26QHH167 pKa = 6.01IVLHH171 pKa = 5.18SVKK174 pKa = 10.65LSGDD178 pKa = 3.82CNISQLNSDD187 pKa = 3.37EE188 pKa = 4.35TFNVNLEE195 pKa = 4.3TTEE198 pKa = 3.95KK199 pKa = 10.85LIGVTVKK206 pKa = 10.63QMGQLDD212 pKa = 4.04SKK214 pKa = 10.31VQSFTGTFQYY224 pKa = 10.83QLSEE228 pKa = 4.26DD229 pKa = 4.01SLDD232 pKa = 3.53GFHH235 pKa = 7.21YY236 pKa = 9.39LICGDD241 pKa = 4.42KK242 pKa = 10.48FFHH245 pKa = 6.88LPNVDD250 pKa = 3.14NGLRR254 pKa = 11.84RR255 pKa = 11.84CVHH258 pKa = 7.01KK259 pKa = 11.21YY260 pKa = 9.73QDD262 pKa = 4.21NKK264 pKa = 9.93TMLYY268 pKa = 9.01TCVNFYY274 pKa = 8.39WLKK277 pKa = 8.82WAFLFFLIMMPITWIMNKK295 pKa = 8.77TKK297 pKa = 10.69GPLSLWYY304 pKa = 9.74DD305 pKa = 3.75LLSIVLYY312 pKa = 9.51PLIYY316 pKa = 10.55LLNNLWSKK324 pKa = 9.82MPYY327 pKa = 9.1RR328 pKa = 11.84CDD330 pKa = 2.98MCGYY334 pKa = 10.67LSLLSHH340 pKa = 7.47DD341 pKa = 4.91CPPKK345 pKa = 10.07CVCGRR350 pKa = 11.84SDD352 pKa = 4.59SGALHH357 pKa = 6.99NPDD360 pKa = 4.35CYY362 pKa = 10.83FNRR365 pKa = 11.84GFPKK369 pKa = 10.39KK370 pKa = 9.63PSKK373 pKa = 10.57KK374 pKa = 10.07FQFLMNTGITRR385 pKa = 11.84NVTFYY390 pKa = 10.04LTRR393 pKa = 11.84FMLSTIVISILPNTLALQASCGYY416 pKa = 10.08NCYY419 pKa = 10.82ADD421 pKa = 5.08LEE423 pKa = 4.42QSKK426 pKa = 10.0FLYY429 pKa = 9.98IGSHH433 pKa = 5.36SMPSARR439 pKa = 11.84EE440 pKa = 3.91CDD442 pKa = 3.59CKK444 pKa = 10.01ITKK447 pKa = 10.33DD448 pKa = 3.55SGSLDD453 pKa = 3.53FPFKK457 pKa = 10.85VVEE460 pKa = 4.43TYY462 pKa = 11.29KK463 pKa = 10.96DD464 pKa = 3.3ITGKK468 pKa = 8.02TQTNIANKK476 pKa = 9.8KK477 pKa = 9.5CYY479 pKa = 9.61PDD481 pKa = 5.04FDD483 pKa = 5.89DD484 pKa = 5.96DD485 pKa = 5.13CADD488 pKa = 3.77HH489 pKa = 7.39LDD491 pKa = 4.0TEE493 pKa = 4.51VLRR496 pKa = 11.84ILACAYY502 pKa = 10.27SCEE505 pKa = 3.92ALKK508 pKa = 10.2MYY510 pKa = 9.94IYY512 pKa = 10.54EE513 pKa = 4.46PLPSVHH519 pKa = 6.58EE520 pKa = 4.49SYY522 pKa = 10.68KK523 pKa = 10.81GEE525 pKa = 4.38AYY527 pKa = 10.6GKK529 pKa = 8.49MKK531 pKa = 10.47PIDD534 pKa = 4.32LYY536 pKa = 11.42SLQKK540 pKa = 10.69SRR542 pKa = 11.84TGVIDD547 pKa = 3.09NRR549 pKa = 11.84TILSRR554 pKa = 11.84QEE556 pKa = 4.0PSLSTWEE563 pKa = 4.08KK564 pKa = 9.09TNSVMSKK571 pKa = 10.62AIASLDD577 pKa = 3.61KK578 pKa = 11.07LDD580 pKa = 3.87VSSISDD586 pKa = 3.7SNTLSRR592 pKa = 11.84TTLMYY597 pKa = 10.41SSKK600 pKa = 10.69VDD602 pKa = 3.19GKK604 pKa = 9.28YY605 pKa = 10.28RR606 pKa = 11.84YY607 pKa = 9.32MIEE610 pKa = 3.75QDD612 pKa = 4.5LISDD616 pKa = 4.15TGTVFEE622 pKa = 4.85MVDD625 pKa = 3.24GTMSHH630 pKa = 6.04SKK632 pKa = 10.93KK633 pKa = 10.94LMITHH638 pKa = 6.74KK639 pKa = 10.51NVGVTYY645 pKa = 9.4TIEE648 pKa = 4.01YY649 pKa = 9.92LYY651 pKa = 8.39TTADD655 pKa = 2.85IVTTKK660 pKa = 10.25TSIYY664 pKa = 10.27SSCTGNCEE672 pKa = 4.16KK673 pKa = 10.48CWEE676 pKa = 4.39GLGSTDD682 pKa = 2.96NPSYY686 pKa = 11.09HH687 pKa = 6.68KK688 pKa = 10.62FCLEE692 pKa = 3.85PTSQWGCEE700 pKa = 3.94DD701 pKa = 3.9FMCIAMNEE709 pKa = 4.14GSTCGHH715 pKa = 7.1CDD717 pKa = 3.06NVYY720 pKa = 11.44DD721 pKa = 4.29MSNSLDD727 pKa = 3.47VYY729 pKa = 9.88RR730 pKa = 11.84VKK732 pKa = 10.43EE733 pKa = 3.88VHH735 pKa = 6.32VSSTACVNAGKK746 pKa = 10.6GYY748 pKa = 10.65NCFNHH753 pKa = 7.67DD754 pKa = 4.28DD755 pKa = 3.82RR756 pKa = 11.84KK757 pKa = 10.95AYY759 pKa = 9.75TNDD762 pKa = 3.14NYY764 pKa = 10.6QVSITSTLQNDD775 pKa = 4.66YY776 pKa = 11.0ISVGHH781 pKa = 6.74KK782 pKa = 10.06LALAKK787 pKa = 10.71DD788 pKa = 4.17GSAYY792 pKa = 9.42TGNIADD798 pKa = 4.91LMSADD803 pKa = 3.69MTFGHH808 pKa = 6.59PQLNKK813 pKa = 10.11QGEE816 pKa = 4.28LLFAEE821 pKa = 4.93EE822 pKa = 4.63YY823 pKa = 9.98IGKK826 pKa = 8.58EE827 pKa = 3.75QLSWDD832 pKa = 4.18CEE834 pKa = 4.31FIGPKK839 pKa = 9.53KK840 pKa = 10.89VYY842 pKa = 9.54IKK844 pKa = 10.75KK845 pKa = 10.26CGFYY849 pKa = 10.39TFDD852 pKa = 3.89HH853 pKa = 6.65FSGLEE858 pKa = 3.76KK859 pKa = 10.72LNEE862 pKa = 4.01GSFVITGLNKK872 pKa = 8.67ITMLKK877 pKa = 10.25DD878 pKa = 3.25YY879 pKa = 10.91SLGKK883 pKa = 10.18IKK885 pKa = 9.72TVLDD889 pKa = 3.74LPIEE893 pKa = 4.21LFVEE897 pKa = 4.4TTEE900 pKa = 4.08APKK903 pKa = 11.11VNIIDD908 pKa = 4.29HH909 pKa = 6.24FCSGCLEE916 pKa = 4.28CFKK919 pKa = 11.28GLDD922 pKa = 3.66CKK924 pKa = 11.31YY925 pKa = 10.44KK926 pKa = 10.49INSNKK931 pKa = 9.57HH932 pKa = 3.82FASRR936 pKa = 11.84ISSDD940 pKa = 3.46FCVSKK945 pKa = 10.24HH946 pKa = 5.31QYY948 pKa = 10.65LIVEE952 pKa = 4.7RR953 pKa = 11.84GSNEE957 pKa = 2.98ISMNLHH963 pKa = 6.33CGVLPEE969 pKa = 3.99NAVLKK974 pKa = 10.01MIVEE978 pKa = 4.44DD979 pKa = 4.2NEE981 pKa = 4.52NISLDD986 pKa = 3.37IPLTKK991 pKa = 9.67IEE993 pKa = 4.98IINDD997 pKa = 3.52KK998 pKa = 10.83NVEE1001 pKa = 4.13TLKK1004 pKa = 10.73DD1005 pKa = 3.33HH1006 pKa = 7.68RR1007 pKa = 11.84IQKK1010 pKa = 9.67SFQHH1014 pKa = 6.9KK1015 pKa = 10.11EE1016 pKa = 3.67DD1017 pKa = 3.85TDD1019 pKa = 3.76FHH1021 pKa = 6.46TFLEE1025 pKa = 4.86TVTYY1029 pKa = 9.07PFNWVANFTNSALDD1043 pKa = 3.9LLKK1046 pKa = 10.81LALLFGFIILGLYY1059 pKa = 9.45VLSKK1063 pKa = 10.6CVNLGKK1069 pKa = 9.94EE1070 pKa = 4.44AYY1072 pKa = 9.26SHH1074 pKa = 5.29ATIYY1078 pKa = 10.27IKK1080 pKa = 10.53KK1081 pKa = 9.83KK1082 pKa = 10.2KK1083 pKa = 9.85HH1084 pKa = 6.8DD1085 pKa = 4.91DD1086 pKa = 3.56NDD1088 pKa = 4.23SDD1090 pKa = 4.21VDD1092 pKa = 3.76DD1093 pKa = 4.63SEE1095 pKa = 4.92MYY1097 pKa = 10.71GEE1099 pKa = 4.33KK1100 pKa = 10.71NLVRR1104 pKa = 11.84RR1105 pKa = 11.84LKK1107 pKa = 11.11YY1108 pKa = 9.83IDD1110 pKa = 3.53LL1111 pKa = 4.45
Molecular weight: 126.28 kDa
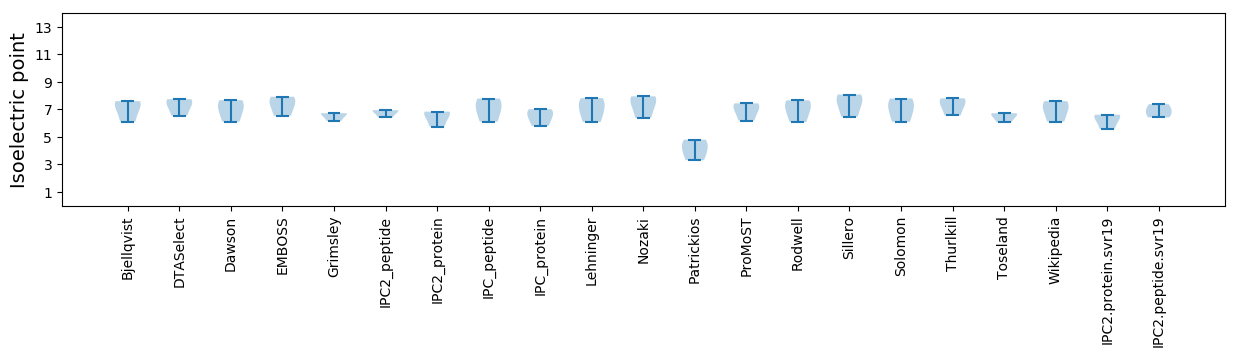
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D4D8M7|A0A0D4D8M7_9VIRU Envelopment polyprotein OS=Groundnut chlorotic fan-spot virus OX=1629551 PE=4 SV=1
MM1 pKa = 7.93DD2 pKa = 5.21MISRR6 pKa = 11.84IGSSINSTVTNGLQNLGRR24 pKa = 11.84TNEE27 pKa = 4.1DD28 pKa = 3.06TDD30 pKa = 4.11VLGSDD35 pKa = 3.37NRR37 pKa = 11.84TLMRR41 pKa = 11.84MPTKK45 pKa = 10.26RR46 pKa = 11.84DD47 pKa = 3.14AEE49 pKa = 4.34KK50 pKa = 10.85ARR52 pKa = 11.84EE53 pKa = 3.81TRR55 pKa = 11.84LQGFKK60 pKa = 10.09EE61 pKa = 4.06RR62 pKa = 11.84NVIEE66 pKa = 4.58SGPSEE71 pKa = 4.0LGEE74 pKa = 3.88YY75 pKa = 10.32DD76 pKa = 3.9GNPVITSDD84 pKa = 4.21LSILEE89 pKa = 3.99RR90 pKa = 11.84LEE92 pKa = 4.65VNTSNHH98 pKa = 4.74ISNWKK103 pKa = 8.3TDD105 pKa = 3.17VFLGNGQEE113 pKa = 4.14MVKK116 pKa = 10.46KK117 pKa = 10.21DD118 pKa = 3.31INLIPTWDD126 pKa = 3.54SKK128 pKa = 11.32KK129 pKa = 10.88EE130 pKa = 3.71YY131 pKa = 8.67MQISRR136 pKa = 11.84VIIWIVPVAPGTKK149 pKa = 10.0GKK151 pKa = 10.25IKK153 pKa = 10.51AALVDD158 pKa = 3.9RR159 pKa = 11.84NKK161 pKa = 10.95AEE163 pKa = 3.9SEE165 pKa = 3.89QIIFQKK171 pKa = 10.63EE172 pKa = 4.09GVLTDD177 pKa = 3.7PLCFIFYY184 pKa = 7.23MHH186 pKa = 7.19WSYY189 pKa = 11.47LKK191 pKa = 10.66SVNNKK196 pKa = 9.12ALCPQLKK203 pKa = 9.9FISNEE208 pKa = 3.98VYY210 pKa = 10.42KK211 pKa = 11.01KK212 pKa = 10.27NVPFAVANFAWRR224 pKa = 11.84KK225 pKa = 7.36VFCNSPIAMTEE236 pKa = 3.92VQPDD240 pKa = 4.58LIVLNRR246 pKa = 11.84GITIRR251 pKa = 11.84NKK253 pKa = 10.69ALLEE257 pKa = 3.97AVKK260 pKa = 10.77CIVPNGNNGKK270 pKa = 7.83TIKK273 pKa = 10.38KK274 pKa = 9.99QIEE277 pKa = 4.37SLSKK281 pKa = 10.57SLEE284 pKa = 3.91AAALEE289 pKa = 4.71DD290 pKa = 4.07EE291 pKa = 4.78SEE293 pKa = 4.36STGAEE298 pKa = 4.02MKK300 pKa = 10.59EE301 pKa = 3.94LTFDD305 pKa = 3.89LL306 pKa = 4.9
MM1 pKa = 7.93DD2 pKa = 5.21MISRR6 pKa = 11.84IGSSINSTVTNGLQNLGRR24 pKa = 11.84TNEE27 pKa = 4.1DD28 pKa = 3.06TDD30 pKa = 4.11VLGSDD35 pKa = 3.37NRR37 pKa = 11.84TLMRR41 pKa = 11.84MPTKK45 pKa = 10.26RR46 pKa = 11.84DD47 pKa = 3.14AEE49 pKa = 4.34KK50 pKa = 10.85ARR52 pKa = 11.84EE53 pKa = 3.81TRR55 pKa = 11.84LQGFKK60 pKa = 10.09EE61 pKa = 4.06RR62 pKa = 11.84NVIEE66 pKa = 4.58SGPSEE71 pKa = 4.0LGEE74 pKa = 3.88YY75 pKa = 10.32DD76 pKa = 3.9GNPVITSDD84 pKa = 4.21LSILEE89 pKa = 3.99RR90 pKa = 11.84LEE92 pKa = 4.65VNTSNHH98 pKa = 4.74ISNWKK103 pKa = 8.3TDD105 pKa = 3.17VFLGNGQEE113 pKa = 4.14MVKK116 pKa = 10.46KK117 pKa = 10.21DD118 pKa = 3.31INLIPTWDD126 pKa = 3.54SKK128 pKa = 11.32KK129 pKa = 10.88EE130 pKa = 3.71YY131 pKa = 8.67MQISRR136 pKa = 11.84VIIWIVPVAPGTKK149 pKa = 10.0GKK151 pKa = 10.25IKK153 pKa = 10.51AALVDD158 pKa = 3.9RR159 pKa = 11.84NKK161 pKa = 10.95AEE163 pKa = 3.9SEE165 pKa = 3.89QIIFQKK171 pKa = 10.63EE172 pKa = 4.09GVLTDD177 pKa = 3.7PLCFIFYY184 pKa = 7.23MHH186 pKa = 7.19WSYY189 pKa = 11.47LKK191 pKa = 10.66SVNNKK196 pKa = 9.12ALCPQLKK203 pKa = 9.9FISNEE208 pKa = 3.98VYY210 pKa = 10.42KK211 pKa = 11.01KK212 pKa = 10.27NVPFAVANFAWRR224 pKa = 11.84KK225 pKa = 7.36VFCNSPIAMTEE236 pKa = 3.92VQPDD240 pKa = 4.58LIVLNRR246 pKa = 11.84GITIRR251 pKa = 11.84NKK253 pKa = 10.69ALLEE257 pKa = 3.97AVKK260 pKa = 10.77CIVPNGNNGKK270 pKa = 7.83TIKK273 pKa = 10.38KK274 pKa = 9.99QIEE277 pKa = 4.37SLSKK281 pKa = 10.57SLEE284 pKa = 3.91AAALEE289 pKa = 4.71DD290 pKa = 4.07EE291 pKa = 4.78SEE293 pKa = 4.36STGAEE298 pKa = 4.02MKK300 pKa = 10.59EE301 pKa = 3.94LTFDD305 pKa = 3.89LL306 pKa = 4.9
Molecular weight: 34.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4287 |
306 |
2870 |
1429.0 |
164.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.732 ± 0.393 | 2.379 ± 0.552 |
6.181 ± 0.256 | 6.461 ± 0.503 |
4.712 ± 0.34 | 4.129 ± 0.65 |
2.029 ± 0.31 | 7.488 ± 0.502 |
8.771 ± 0.264 | 9.447 ± 0.302 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.732 ± 0.367 | 6.765 ± 0.374 |
2.892 ± 0.269 | 2.426 ± 0.108 |
3.569 ± 0.519 | 9.097 ± 0.312 |
5.482 ± 0.485 | 5.365 ± 0.333 |
0.77 ± 0.204 | 4.572 ± 0.598 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
