
Epulopiscium sp. SCG-C07WGA-EpuloA2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Epulopiscium; unclassified Epulopiscium
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
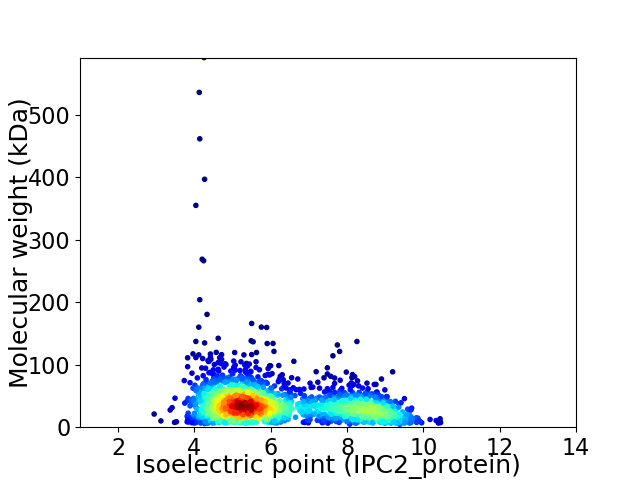
Virtual 2D-PAGE plot for 1923 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V2MA43|A0A1V2MA43_9FIRM Pyridoxal phosphate homeostasis protein OS=Epulopiscium sp. SCG-C07WGA-EpuloA2 OX=1712378 GN=AN641_00675 PE=3 SV=1
MM1 pKa = 7.59KK2 pKa = 10.44KK3 pKa = 10.03FLSVVLSSTMLIAFAGCTSSEE24 pKa = 4.56TNTSTDD30 pKa = 2.62TSNLVGVSMPTQDD43 pKa = 3.63LQRR46 pKa = 11.84WNQDD50 pKa = 3.07GEE52 pKa = 4.23NMKK55 pKa = 10.72AQLEE59 pKa = 4.46SAGYY63 pKa = 10.35DD64 pKa = 2.9VDD66 pKa = 4.09LQYY69 pKa = 11.57AANDD73 pKa = 3.1IATQVSQIEE82 pKa = 4.11NMISGGVKK90 pKa = 8.58TLVVASIDD98 pKa = 3.6GSALGTVMEE107 pKa = 4.26QAKK110 pKa = 10.84SNGIEE115 pKa = 3.99VIAYY119 pKa = 8.34DD120 pKa = 4.26RR121 pKa = 11.84LIMGTDD127 pKa = 3.94AISYY131 pKa = 10.43YY132 pKa = 9.8STFDD136 pKa = 3.08NWDD139 pKa = 3.33VGVKK143 pKa = 9.18QGEE146 pKa = 4.53YY147 pKa = 10.15IVDD150 pKa = 3.73KK151 pKa = 11.14LKK153 pKa = 10.95LDD155 pKa = 4.09STDD158 pKa = 2.91EE159 pKa = 4.3TYY161 pKa = 10.37TIEE164 pKa = 5.96FITGDD169 pKa = 3.84PGDD172 pKa = 3.73NNINFFFGGAMSILQPYY189 pKa = 9.5LDD191 pKa = 4.77EE192 pKa = 4.79GTLEE196 pKa = 4.22CLSGQTEE203 pKa = 4.12QTEE206 pKa = 4.47VATVNWATDD215 pKa = 3.25VAQARR220 pKa = 11.84FEE222 pKa = 4.84NILASYY228 pKa = 8.34YY229 pKa = 10.51TDD231 pKa = 3.29TDD233 pKa = 3.9LDD235 pKa = 3.62IVLASNDD242 pKa = 3.13STAQGVTNALVSSYY256 pKa = 11.38DD257 pKa = 3.12GAYY260 pKa = 9.81PLLTGQDD267 pKa = 3.59CDD269 pKa = 3.72VVSVQNIISGKK280 pKa = 8.2QEE282 pKa = 3.4MSIFKK287 pKa = 9.0DD288 pKa = 3.49TRR290 pKa = 11.84DD291 pKa = 3.43LAAKK295 pKa = 8.05TVEE298 pKa = 4.23MVDD301 pKa = 3.8ALMKK305 pKa = 10.69GSEE308 pKa = 4.32PPINDD313 pKa = 2.81NKK315 pKa = 10.8TYY317 pKa = 11.18DD318 pKa = 3.73NGTGIIPSYY327 pKa = 10.14LCEE330 pKa = 4.08PVVATLNNYY339 pKa = 7.54EE340 pKa = 4.28ALLIEE345 pKa = 4.33TGYY348 pKa = 8.74YY349 pKa = 8.05TAEE352 pKa = 4.04EE353 pKa = 4.28VQPP356 pKa = 4.05
MM1 pKa = 7.59KK2 pKa = 10.44KK3 pKa = 10.03FLSVVLSSTMLIAFAGCTSSEE24 pKa = 4.56TNTSTDD30 pKa = 2.62TSNLVGVSMPTQDD43 pKa = 3.63LQRR46 pKa = 11.84WNQDD50 pKa = 3.07GEE52 pKa = 4.23NMKK55 pKa = 10.72AQLEE59 pKa = 4.46SAGYY63 pKa = 10.35DD64 pKa = 2.9VDD66 pKa = 4.09LQYY69 pKa = 11.57AANDD73 pKa = 3.1IATQVSQIEE82 pKa = 4.11NMISGGVKK90 pKa = 8.58TLVVASIDD98 pKa = 3.6GSALGTVMEE107 pKa = 4.26QAKK110 pKa = 10.84SNGIEE115 pKa = 3.99VIAYY119 pKa = 8.34DD120 pKa = 4.26RR121 pKa = 11.84LIMGTDD127 pKa = 3.94AISYY131 pKa = 10.43YY132 pKa = 9.8STFDD136 pKa = 3.08NWDD139 pKa = 3.33VGVKK143 pKa = 9.18QGEE146 pKa = 4.53YY147 pKa = 10.15IVDD150 pKa = 3.73KK151 pKa = 11.14LKK153 pKa = 10.95LDD155 pKa = 4.09STDD158 pKa = 2.91EE159 pKa = 4.3TYY161 pKa = 10.37TIEE164 pKa = 5.96FITGDD169 pKa = 3.84PGDD172 pKa = 3.73NNINFFFGGAMSILQPYY189 pKa = 9.5LDD191 pKa = 4.77EE192 pKa = 4.79GTLEE196 pKa = 4.22CLSGQTEE203 pKa = 4.12QTEE206 pKa = 4.47VATVNWATDD215 pKa = 3.25VAQARR220 pKa = 11.84FEE222 pKa = 4.84NILASYY228 pKa = 8.34YY229 pKa = 10.51TDD231 pKa = 3.29TDD233 pKa = 3.9LDD235 pKa = 3.62IVLASNDD242 pKa = 3.13STAQGVTNALVSSYY256 pKa = 11.38DD257 pKa = 3.12GAYY260 pKa = 9.81PLLTGQDD267 pKa = 3.59CDD269 pKa = 3.72VVSVQNIISGKK280 pKa = 8.2QEE282 pKa = 3.4MSIFKK287 pKa = 9.0DD288 pKa = 3.49TRR290 pKa = 11.84DD291 pKa = 3.43LAAKK295 pKa = 8.05TVEE298 pKa = 4.23MVDD301 pKa = 3.8ALMKK305 pKa = 10.69GSEE308 pKa = 4.32PPINDD313 pKa = 2.81NKK315 pKa = 10.8TYY317 pKa = 11.18DD318 pKa = 3.73NGTGIIPSYY327 pKa = 10.14LCEE330 pKa = 4.08PVVATLNNYY339 pKa = 7.54EE340 pKa = 4.28ALLIEE345 pKa = 4.33TGYY348 pKa = 8.74YY349 pKa = 8.05TAEE352 pKa = 4.04EE353 pKa = 4.28VQPP356 pKa = 4.05
Molecular weight: 38.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V2MB14|A0A1V2MB14_9FIRM Coenzyme A biosynthesis bifunctional protein CoaBC OS=Epulopiscium sp. SCG-C07WGA-EpuloA2 OX=1712378 GN=coaBC PE=3 SV=1
MM1 pKa = 7.2KK2 pKa = 10.24RR3 pKa = 11.84VFAFPLYY10 pKa = 10.36LLSIQFSCHH19 pKa = 5.0SCPFQAATAKK29 pKa = 10.06VAEE32 pKa = 4.32LRR34 pKa = 11.84RR35 pKa = 11.84TIVNKK40 pKa = 7.05QTKK43 pKa = 8.5LHH45 pKa = 5.81PRR47 pKa = 11.84KK48 pKa = 10.11VFLGSPLRR56 pKa = 11.84IFWVGDD62 pKa = 3.56STEE65 pKa = 4.73LEE67 pKa = 4.25AKK69 pKa = 9.96SGRR72 pKa = 11.84SLLSQTGAGGLVGPGRR88 pKa = 11.84GGHH91 pKa = 4.36WRR93 pKa = 11.84GGKK96 pKa = 9.22VRR98 pKa = 11.84GRR100 pKa = 11.84RR101 pKa = 11.84GKK103 pKa = 9.25GTFAVQLAALCEE115 pKa = 4.15AGTVGRR121 pKa = 11.84YY122 pKa = 8.37RR123 pKa = 11.84AGEE126 pKa = 3.83EE127 pKa = 4.24SEE129 pKa = 4.16
MM1 pKa = 7.2KK2 pKa = 10.24RR3 pKa = 11.84VFAFPLYY10 pKa = 10.36LLSIQFSCHH19 pKa = 5.0SCPFQAATAKK29 pKa = 10.06VAEE32 pKa = 4.32LRR34 pKa = 11.84RR35 pKa = 11.84TIVNKK40 pKa = 7.05QTKK43 pKa = 8.5LHH45 pKa = 5.81PRR47 pKa = 11.84KK48 pKa = 10.11VFLGSPLRR56 pKa = 11.84IFWVGDD62 pKa = 3.56STEE65 pKa = 4.73LEE67 pKa = 4.25AKK69 pKa = 9.96SGRR72 pKa = 11.84SLLSQTGAGGLVGPGRR88 pKa = 11.84GGHH91 pKa = 4.36WRR93 pKa = 11.84GGKK96 pKa = 9.22VRR98 pKa = 11.84GRR100 pKa = 11.84RR101 pKa = 11.84GKK103 pKa = 9.25GTFAVQLAALCEE115 pKa = 4.15AGTVGRR121 pKa = 11.84YY122 pKa = 8.37RR123 pKa = 11.84AGEE126 pKa = 3.83EE127 pKa = 4.24SEE129 pKa = 4.16
Molecular weight: 13.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
670132 |
37 |
5516 |
348.5 |
39.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.196 ± 0.068 | 1.061 ± 0.023 |
5.712 ± 0.063 | 7.165 ± 0.066 |
4.433 ± 0.053 | 6.069 ± 0.053 |
1.47 ± 0.023 | 10.135 ± 0.075 |
7.743 ± 0.064 | 8.962 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.484 ± 0.032 | 6.344 ± 0.054 |
3.16 ± 0.033 | 3.197 ± 0.032 |
2.848 ± 0.038 | 5.949 ± 0.044 |
6.06 ± 0.101 | 6.092 ± 0.047 |
0.785 ± 0.017 | 4.136 ± 0.044 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
