
Halolamina pelagica
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halolamina
Average proteome isoelectric point is 5.09
Get precalculated fractions of proteins
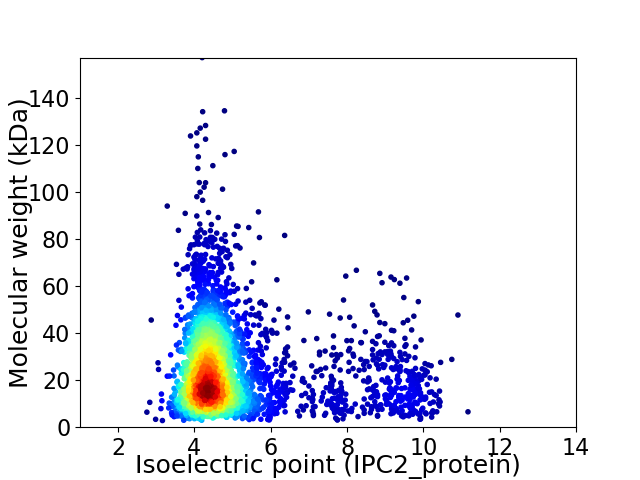
Virtual 2D-PAGE plot for 3464 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N8HZJ8|A0A0N8HZJ8_9EURY tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS OS=Halolamina pelagica OX=699431 GN=tiaS_1 PE=4 SV=1
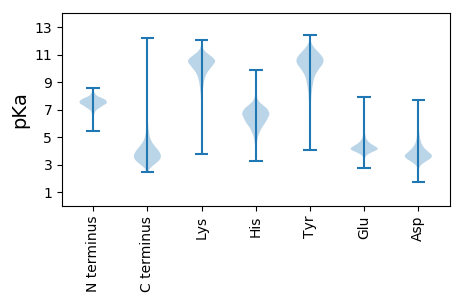
MM1 pKa = 7.13TGPWAQWDD9 pKa = 4.0HH10 pKa = 5.13VLKK13 pKa = 10.77VDD15 pKa = 4.28PDD17 pKa = 3.82KK18 pKa = 11.45EE19 pKa = 4.13LLDD22 pKa = 5.48DD23 pKa = 3.81EE24 pKa = 5.11TFDD27 pKa = 5.25DD28 pKa = 4.5VAATGTDD35 pKa = 3.09ALEE38 pKa = 4.31IGGTMDD44 pKa = 3.22VTSEE48 pKa = 3.54KK49 pKa = 9.45MARR52 pKa = 11.84VVDD55 pKa = 4.03ACAAHH60 pKa = 6.99DD61 pKa = 3.74VPIYY65 pKa = 10.11QEE67 pKa = 3.88PSNPGVVIDD76 pKa = 5.06HH77 pKa = 6.84EE78 pKa = 4.56ALDD81 pKa = 4.95GYY83 pKa = 10.49LVPTVLNAGDD93 pKa = 4.26PFWITGAHH101 pKa = 6.22KK102 pKa = 9.33EE103 pKa = 3.86WARR106 pKa = 11.84IDD108 pKa = 5.82DD109 pKa = 4.47DD110 pKa = 5.66LDD112 pKa = 3.3WANTHH117 pKa = 5.25TEE119 pKa = 4.15AYY121 pKa = 10.2VVLNPDD127 pKa = 3.37AAVAEE132 pKa = 4.35YY133 pKa = 10.65TEE135 pKa = 4.81ADD137 pKa = 3.69CEE139 pKa = 4.27LTAEE143 pKa = 4.36EE144 pKa = 4.23VAAYY148 pKa = 10.24AGVAEE153 pKa = 4.72RR154 pKa = 11.84MFGQDD159 pKa = 2.44IVYY162 pKa = 10.51LEE164 pKa = 4.01YY165 pKa = 10.88SGTYY169 pKa = 10.14GDD171 pKa = 4.77PEE173 pKa = 4.4TVAAAADD180 pKa = 3.98EE181 pKa = 5.06LDD183 pKa = 4.47DD184 pKa = 3.83ATLFYY189 pKa = 11.24GGGIHH194 pKa = 7.56DD195 pKa = 4.3YY196 pKa = 10.92EE197 pKa = 4.79SARR200 pKa = 11.84EE201 pKa = 4.03MGEE204 pKa = 3.85HH205 pKa = 7.19ADD207 pKa = 3.82TVVVGDD213 pKa = 4.37LLHH216 pKa = 7.44DD217 pKa = 4.24EE218 pKa = 4.95GVDD221 pKa = 3.71AVAEE225 pKa = 4.36TVDD228 pKa = 3.65GAKK231 pKa = 10.47DD232 pKa = 3.13AA233 pKa = 4.74
MM1 pKa = 7.13TGPWAQWDD9 pKa = 4.0HH10 pKa = 5.13VLKK13 pKa = 10.77VDD15 pKa = 4.28PDD17 pKa = 3.82KK18 pKa = 11.45EE19 pKa = 4.13LLDD22 pKa = 5.48DD23 pKa = 3.81EE24 pKa = 5.11TFDD27 pKa = 5.25DD28 pKa = 4.5VAATGTDD35 pKa = 3.09ALEE38 pKa = 4.31IGGTMDD44 pKa = 3.22VTSEE48 pKa = 3.54KK49 pKa = 9.45MARR52 pKa = 11.84VVDD55 pKa = 4.03ACAAHH60 pKa = 6.99DD61 pKa = 3.74VPIYY65 pKa = 10.11QEE67 pKa = 3.88PSNPGVVIDD76 pKa = 5.06HH77 pKa = 6.84EE78 pKa = 4.56ALDD81 pKa = 4.95GYY83 pKa = 10.49LVPTVLNAGDD93 pKa = 4.26PFWITGAHH101 pKa = 6.22KK102 pKa = 9.33EE103 pKa = 3.86WARR106 pKa = 11.84IDD108 pKa = 5.82DD109 pKa = 4.47DD110 pKa = 5.66LDD112 pKa = 3.3WANTHH117 pKa = 5.25TEE119 pKa = 4.15AYY121 pKa = 10.2VVLNPDD127 pKa = 3.37AAVAEE132 pKa = 4.35YY133 pKa = 10.65TEE135 pKa = 4.81ADD137 pKa = 3.69CEE139 pKa = 4.27LTAEE143 pKa = 4.36EE144 pKa = 4.23VAAYY148 pKa = 10.24AGVAEE153 pKa = 4.72RR154 pKa = 11.84MFGQDD159 pKa = 2.44IVYY162 pKa = 10.51LEE164 pKa = 4.01YY165 pKa = 10.88SGTYY169 pKa = 10.14GDD171 pKa = 4.77PEE173 pKa = 4.4TVAAAADD180 pKa = 3.98EE181 pKa = 5.06LDD183 pKa = 4.47DD184 pKa = 3.83ATLFYY189 pKa = 11.24GGGIHH194 pKa = 7.56DD195 pKa = 4.3YY196 pKa = 10.92EE197 pKa = 4.79SARR200 pKa = 11.84EE201 pKa = 4.03MGEE204 pKa = 3.85HH205 pKa = 7.19ADD207 pKa = 3.82TVVVGDD213 pKa = 4.37LLHH216 pKa = 7.44DD217 pKa = 4.24EE218 pKa = 4.95GVDD221 pKa = 3.71AVAEE225 pKa = 4.36TVDD228 pKa = 3.65GAKK231 pKa = 10.47DD232 pKa = 3.13AA233 pKa = 4.74
Molecular weight: 25.07 kDa
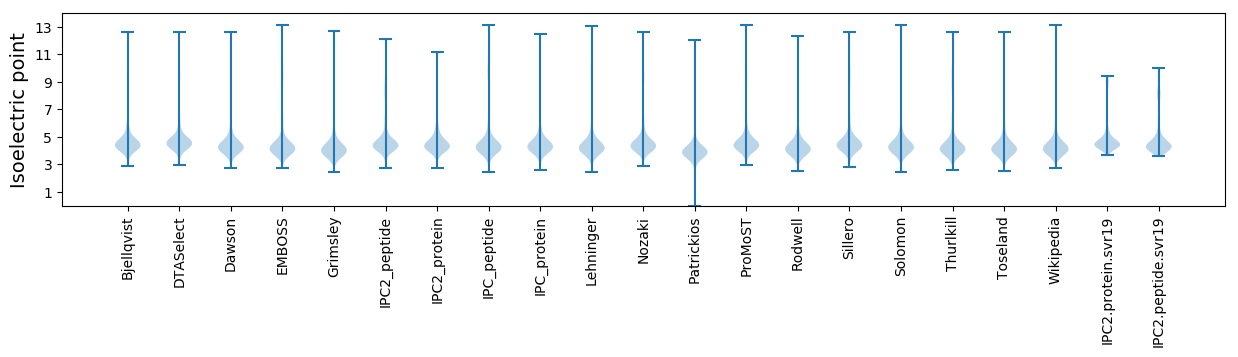
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P7GV22|A0A0P7GV22_9EURY Uncharacterized protein OS=Halolamina pelagica OX=699431 GN=SY89_00086 PE=4 SV=1
MM1 pKa = 7.27ARR3 pKa = 11.84KK4 pKa = 9.04SVRR7 pKa = 11.84VRR9 pKa = 11.84KK10 pKa = 9.71VRR12 pKa = 11.84YY13 pKa = 8.58KK14 pKa = 10.04GHH16 pKa = 4.63TVRR19 pKa = 11.84VKK21 pKa = 9.34VYY23 pKa = 9.36RR24 pKa = 11.84RR25 pKa = 11.84NGRR28 pKa = 11.84IVRR31 pKa = 11.84LKK33 pKa = 10.43VIKK36 pKa = 10.81SPVKK40 pKa = 9.86VKK42 pKa = 10.61RR43 pKa = 11.84KK44 pKa = 9.17VLIRR48 pKa = 11.84RR49 pKa = 11.84IRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 3.26
MM1 pKa = 7.27ARR3 pKa = 11.84KK4 pKa = 9.04SVRR7 pKa = 11.84VRR9 pKa = 11.84KK10 pKa = 9.71VRR12 pKa = 11.84YY13 pKa = 8.58KK14 pKa = 10.04GHH16 pKa = 4.63TVRR19 pKa = 11.84VKK21 pKa = 9.34VYY23 pKa = 9.36RR24 pKa = 11.84RR25 pKa = 11.84NGRR28 pKa = 11.84IVRR31 pKa = 11.84LKK33 pKa = 10.43VIKK36 pKa = 10.81SPVKK40 pKa = 9.86VKK42 pKa = 10.61RR43 pKa = 11.84KK44 pKa = 9.17VLIRR48 pKa = 11.84RR49 pKa = 11.84IRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 3.26
Molecular weight: 6.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
838665 |
29 |
1422 |
242.1 |
26.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.781 ± 0.071 | 0.748 ± 0.016 |
8.054 ± 0.051 | 8.359 ± 0.065 |
3.248 ± 0.03 | 8.73 ± 0.045 |
1.938 ± 0.022 | 3.709 ± 0.033 |
1.668 ± 0.023 | 8.879 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.79 ± 0.017 | 2.174 ± 0.025 |
4.954 ± 0.031 | 2.436 ± 0.025 |
6.861 ± 0.05 | 5.62 ± 0.037 |
6.397 ± 0.039 | 8.917 ± 0.042 |
1.19 ± 0.018 | 2.547 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
