
Microbacterium sp. TNHR37B
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
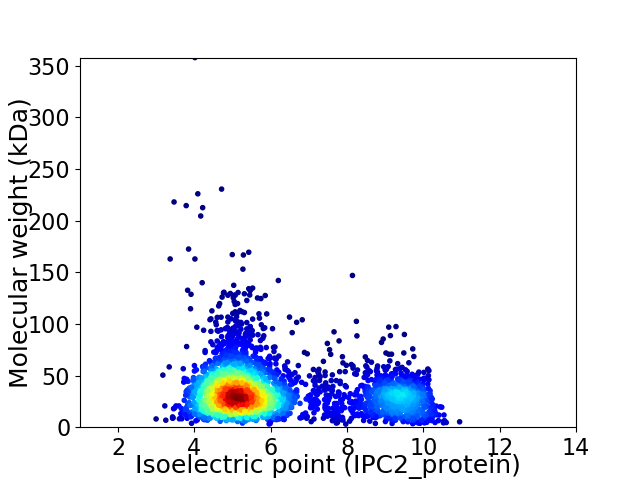
Virtual 2D-PAGE plot for 3221 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A164AXD9|A0A164AXD9_9MICO Bifunctional protein PutA OS=Microbacterium sp. TNHR37B OX=1775956 GN=putA PE=3 SV=1
MM1 pKa = 7.59NKK3 pKa = 9.86RR4 pKa = 11.84LLAAAAAVAATALALSACSAGTAPKK29 pKa = 10.58GDD31 pKa = 3.88DD32 pKa = 4.45AIDD35 pKa = 4.41EE36 pKa = 4.27IRR38 pKa = 11.84VAATDD43 pKa = 3.75PQLIIPGRR51 pKa = 11.84QSVAYY56 pKa = 8.94DD57 pKa = 3.09VNMAVWAPLAFLNSDD72 pKa = 3.36GSLDD76 pKa = 3.77YY77 pKa = 11.24VQAEE81 pKa = 4.53SIEE84 pKa = 4.23SDD86 pKa = 4.57DD87 pKa = 5.26AITWTITLRR96 pKa = 11.84EE97 pKa = 4.02GWTFQDD103 pKa = 3.8GTPVTAQSYY112 pKa = 8.15VDD114 pKa = 3.3SWNAVAYY121 pKa = 9.93GPNAFEE127 pKa = 5.69NSGQLANIVGYY138 pKa = 11.18ADD140 pKa = 4.49LNPAEE145 pKa = 5.03GEE147 pKa = 4.02PSTTEE152 pKa = 3.63MSGLAVVDD160 pKa = 3.68EE161 pKa = 4.42RR162 pKa = 11.84TFTVEE167 pKa = 3.53LTGPDD172 pKa = 3.44SQFPLQVTQAQTAMFPMPASALTDD196 pKa = 3.82FDD198 pKa = 5.13AYY200 pKa = 9.28NTHH203 pKa = 7.62PIGNGPFAFAEE214 pKa = 4.42DD215 pKa = 4.09YY216 pKa = 11.29VEE218 pKa = 4.51NEE220 pKa = 4.4PIVLEE225 pKa = 5.35AYY227 pKa = 10.25DD228 pKa = 4.97DD229 pKa = 4.09YY230 pKa = 11.6QGEE233 pKa = 4.48KK234 pKa = 8.8PTIDD238 pKa = 3.95RR239 pKa = 11.84ITFVPYY245 pKa = 9.76TDD247 pKa = 4.54SEE249 pKa = 4.41TAYY252 pKa = 9.98TDD254 pKa = 3.57VLAGNLDD261 pKa = 3.83VAGVPASKK269 pKa = 9.11LTQAAGDD276 pKa = 3.78FGDD279 pKa = 3.52RR280 pKa = 11.84LYY282 pKa = 11.37SFEE285 pKa = 4.42APGISFLGLPLWNDD299 pKa = 3.1AYY301 pKa = 11.22DD302 pKa = 4.68DD303 pKa = 3.52IRR305 pKa = 11.84VRR307 pKa = 11.84QAISMAIDD315 pKa = 3.3RR316 pKa = 11.84EE317 pKa = 4.3AIIDD321 pKa = 3.89VIYY324 pKa = 10.72GGTYY328 pKa = 10.44DD329 pKa = 5.86AATAWTPAIEE339 pKa = 4.88PGTPAGICGEE349 pKa = 4.08YY350 pKa = 10.46CEE352 pKa = 4.98YY353 pKa = 11.19DD354 pKa = 3.59PEE356 pKa = 4.22AAKK359 pKa = 10.86ALLDD363 pKa = 3.71EE364 pKa = 5.52AGGFDD369 pKa = 4.13GSLEE373 pKa = 4.01IYY375 pKa = 10.43FPGGGGLDD383 pKa = 3.63PLYY386 pKa = 10.66EE387 pKa = 4.88AIANQLRR394 pKa = 11.84QNLGIEE400 pKa = 4.32ATAKK404 pKa = 10.5PSADD408 pKa = 2.68WAEE411 pKa = 4.02FLQNRR416 pKa = 11.84NDD418 pKa = 3.69EE419 pKa = 4.79NIDD422 pKa = 3.47GPFFSRR428 pKa = 11.84WGALYY433 pKa = 9.55PSQQATLRR441 pKa = 11.84VFFIEE446 pKa = 5.79GGGCTNCIPWYY457 pKa = 9.43SDD459 pKa = 3.02EE460 pKa = 4.58VADD463 pKa = 5.73AMAAADD469 pKa = 4.68ADD471 pKa = 4.07LSTDD475 pKa = 3.05GSAYY479 pKa = 10.62AEE481 pKa = 4.17VQKK484 pKa = 10.75LIQAEE489 pKa = 4.83FPAVPLFFEE498 pKa = 4.66KK499 pKa = 10.77YY500 pKa = 10.29SYY502 pKa = 9.07VTSEE506 pKa = 3.98KK507 pKa = 10.76VADD510 pKa = 4.1LVTSPVGNPTYY521 pKa = 10.59RR522 pKa = 11.84SIVLDD527 pKa = 3.96DD528 pKa = 3.64
MM1 pKa = 7.59NKK3 pKa = 9.86RR4 pKa = 11.84LLAAAAAVAATALALSACSAGTAPKK29 pKa = 10.58GDD31 pKa = 3.88DD32 pKa = 4.45AIDD35 pKa = 4.41EE36 pKa = 4.27IRR38 pKa = 11.84VAATDD43 pKa = 3.75PQLIIPGRR51 pKa = 11.84QSVAYY56 pKa = 8.94DD57 pKa = 3.09VNMAVWAPLAFLNSDD72 pKa = 3.36GSLDD76 pKa = 3.77YY77 pKa = 11.24VQAEE81 pKa = 4.53SIEE84 pKa = 4.23SDD86 pKa = 4.57DD87 pKa = 5.26AITWTITLRR96 pKa = 11.84EE97 pKa = 4.02GWTFQDD103 pKa = 3.8GTPVTAQSYY112 pKa = 8.15VDD114 pKa = 3.3SWNAVAYY121 pKa = 9.93GPNAFEE127 pKa = 5.69NSGQLANIVGYY138 pKa = 11.18ADD140 pKa = 4.49LNPAEE145 pKa = 5.03GEE147 pKa = 4.02PSTTEE152 pKa = 3.63MSGLAVVDD160 pKa = 3.68EE161 pKa = 4.42RR162 pKa = 11.84TFTVEE167 pKa = 3.53LTGPDD172 pKa = 3.44SQFPLQVTQAQTAMFPMPASALTDD196 pKa = 3.82FDD198 pKa = 5.13AYY200 pKa = 9.28NTHH203 pKa = 7.62PIGNGPFAFAEE214 pKa = 4.42DD215 pKa = 4.09YY216 pKa = 11.29VEE218 pKa = 4.51NEE220 pKa = 4.4PIVLEE225 pKa = 5.35AYY227 pKa = 10.25DD228 pKa = 4.97DD229 pKa = 4.09YY230 pKa = 11.6QGEE233 pKa = 4.48KK234 pKa = 8.8PTIDD238 pKa = 3.95RR239 pKa = 11.84ITFVPYY245 pKa = 9.76TDD247 pKa = 4.54SEE249 pKa = 4.41TAYY252 pKa = 9.98TDD254 pKa = 3.57VLAGNLDD261 pKa = 3.83VAGVPASKK269 pKa = 9.11LTQAAGDD276 pKa = 3.78FGDD279 pKa = 3.52RR280 pKa = 11.84LYY282 pKa = 11.37SFEE285 pKa = 4.42APGISFLGLPLWNDD299 pKa = 3.1AYY301 pKa = 11.22DD302 pKa = 4.68DD303 pKa = 3.52IRR305 pKa = 11.84VRR307 pKa = 11.84QAISMAIDD315 pKa = 3.3RR316 pKa = 11.84EE317 pKa = 4.3AIIDD321 pKa = 3.89VIYY324 pKa = 10.72GGTYY328 pKa = 10.44DD329 pKa = 5.86AATAWTPAIEE339 pKa = 4.88PGTPAGICGEE349 pKa = 4.08YY350 pKa = 10.46CEE352 pKa = 4.98YY353 pKa = 11.19DD354 pKa = 3.59PEE356 pKa = 4.22AAKK359 pKa = 10.86ALLDD363 pKa = 3.71EE364 pKa = 5.52AGGFDD369 pKa = 4.13GSLEE373 pKa = 4.01IYY375 pKa = 10.43FPGGGGLDD383 pKa = 3.63PLYY386 pKa = 10.66EE387 pKa = 4.88AIANQLRR394 pKa = 11.84QNLGIEE400 pKa = 4.32ATAKK404 pKa = 10.5PSADD408 pKa = 2.68WAEE411 pKa = 4.02FLQNRR416 pKa = 11.84NDD418 pKa = 3.69EE419 pKa = 4.79NIDD422 pKa = 3.47GPFFSRR428 pKa = 11.84WGALYY433 pKa = 9.55PSQQATLRR441 pKa = 11.84VFFIEE446 pKa = 5.79GGGCTNCIPWYY457 pKa = 9.43SDD459 pKa = 3.02EE460 pKa = 4.58VADD463 pKa = 5.73AMAAADD469 pKa = 4.68ADD471 pKa = 4.07LSTDD475 pKa = 3.05GSAYY479 pKa = 10.62AEE481 pKa = 4.17VQKK484 pKa = 10.75LIQAEE489 pKa = 4.83FPAVPLFFEE498 pKa = 4.66KK499 pKa = 10.77YY500 pKa = 10.29SYY502 pKa = 9.07VTSEE506 pKa = 3.98KK507 pKa = 10.76VADD510 pKa = 4.1LVTSPVGNPTYY521 pKa = 10.59RR522 pKa = 11.84SIVLDD527 pKa = 3.96DD528 pKa = 3.64
Molecular weight: 56.6 kDa
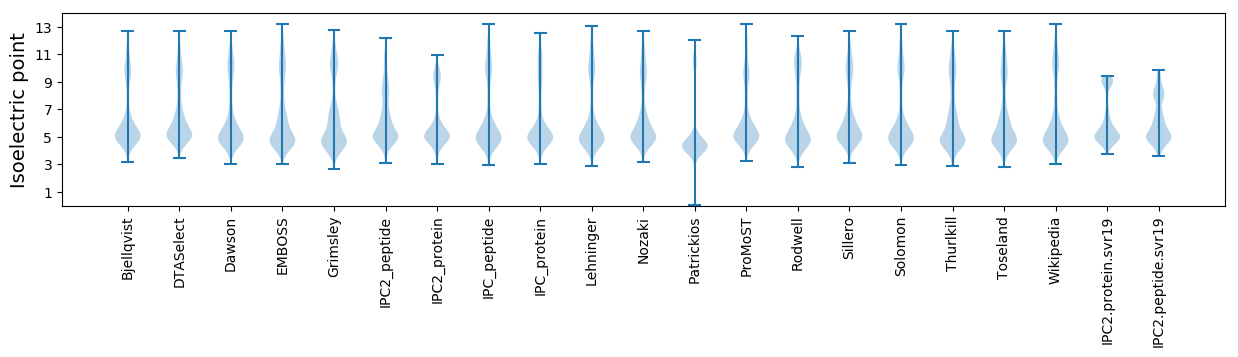
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A164BLB5|A0A164BLB5_9MICO DUF305 domain-containing protein OS=Microbacterium sp. TNHR37B OX=1775956 GN=AVP41_02624 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.64GRR40 pKa = 11.84TEE42 pKa = 4.13LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.64GRR40 pKa = 11.84TEE42 pKa = 4.13LSAA45 pKa = 4.86
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1083291 |
29 |
3542 |
336.3 |
35.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.738 ± 0.057 | 0.482 ± 0.009 |
6.301 ± 0.035 | 5.495 ± 0.037 |
3.063 ± 0.026 | 8.924 ± 0.036 |
1.984 ± 0.023 | 4.298 ± 0.035 |
1.755 ± 0.032 | 10.02 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.664 ± 0.018 | 1.782 ± 0.021 |
5.551 ± 0.029 | 2.626 ± 0.021 |
7.632 ± 0.055 | 5.647 ± 0.034 |
6.303 ± 0.053 | 9.157 ± 0.049 |
1.608 ± 0.019 | 1.97 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
