
Dickeya phage Dagda_B1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Studiervirinae; Aarhusvirus; unclassified Aarhusvirus
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
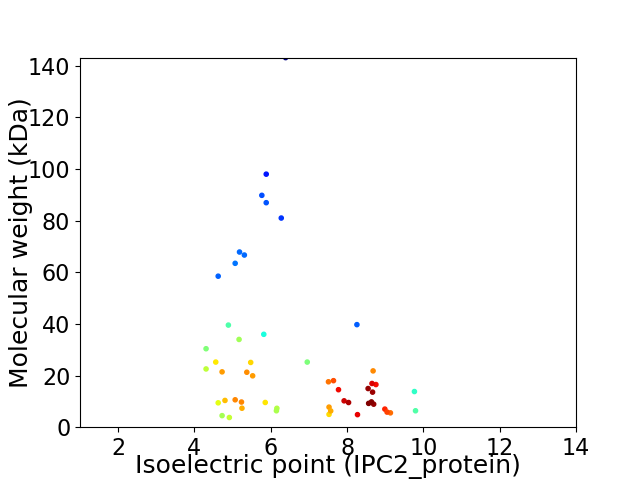
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385IFH9|A0A385IFH9_9CAUD Uncharacterized protein OS=Dickeya phage Dagda_B1 OX=2320189 PE=4 SV=1
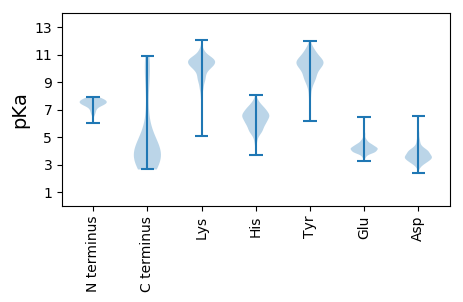
MM1 pKa = 7.24TDD3 pKa = 2.96IYY5 pKa = 11.38AEE7 pKa = 4.12YY8 pKa = 9.99GVTGAVMSEE17 pKa = 3.76PSEE20 pKa = 4.4GYY22 pKa = 9.53NEE24 pKa = 3.96QMIAQPVDD32 pKa = 3.26VRR34 pKa = 11.84DD35 pKa = 3.98GDD37 pKa = 4.09DD38 pKa = 3.95SITVEE43 pKa = 3.97SEE45 pKa = 3.87EE46 pKa = 4.11FEE48 pKa = 4.28EE49 pKa = 5.04ASSEE53 pKa = 3.92EE54 pKa = 4.17LPEE57 pKa = 4.71GEE59 pKa = 4.37EE60 pKa = 3.89QGEE63 pKa = 4.28EE64 pKa = 3.95QSGEE68 pKa = 4.05EE69 pKa = 4.13SGEE72 pKa = 4.14EE73 pKa = 3.66AGEE76 pKa = 3.88AAEE79 pKa = 4.69FEE81 pKa = 4.55AVGDD85 pKa = 3.93VPEE88 pKa = 4.33EE89 pKa = 4.0LVTVTTRR96 pKa = 11.84IAEE99 pKa = 4.23SEE101 pKa = 4.49SAFNDD106 pKa = 3.47MVADD110 pKa = 4.46AAARR114 pKa = 11.84GLDD117 pKa = 3.4QEE119 pKa = 4.47SFAIISAEE127 pKa = 3.92YY128 pKa = 9.0EE129 pKa = 4.19AEE131 pKa = 4.42GEE133 pKa = 4.23LSPKK137 pKa = 10.37SYY139 pKa = 10.51EE140 pKa = 3.93ALAKK144 pKa = 10.39VGYY147 pKa = 9.13SKK149 pKa = 11.31NFIDD153 pKa = 5.67SFIAGQEE160 pKa = 4.36AIGSQYY166 pKa = 10.28MSAIQAYY173 pKa = 9.85AGGEE177 pKa = 4.02AKK179 pKa = 10.3FQAIFAHH186 pKa = 6.37LQASSPEE193 pKa = 4.08SAEE196 pKa = 3.87ALEE199 pKa = 4.14NAIGQRR205 pKa = 11.84DD206 pKa = 3.77LKK208 pKa = 10.67AVKK211 pKa = 10.48GIINLAASSVRR222 pKa = 11.84KK223 pKa = 9.76EE224 pKa = 3.89FGKK227 pKa = 9.7PAEE230 pKa = 4.13RR231 pKa = 11.84TLSKK235 pKa = 10.38RR236 pKa = 11.84ATPAKK241 pKa = 9.08VTAPKK246 pKa = 10.39AAGFEE251 pKa = 4.65SQAEE255 pKa = 4.24MVKK258 pKa = 10.68AMKK261 pKa = 10.08DD262 pKa = 3.28PKK264 pKa = 10.08YY265 pKa = 10.96GRR267 pKa = 11.84DD268 pKa = 3.13AAYY271 pKa = 8.85TRR273 pKa = 11.84SVEE276 pKa = 4.69LKK278 pKa = 10.21VLNAKK283 pKa = 9.48FF284 pKa = 3.3
MM1 pKa = 7.24TDD3 pKa = 2.96IYY5 pKa = 11.38AEE7 pKa = 4.12YY8 pKa = 9.99GVTGAVMSEE17 pKa = 3.76PSEE20 pKa = 4.4GYY22 pKa = 9.53NEE24 pKa = 3.96QMIAQPVDD32 pKa = 3.26VRR34 pKa = 11.84DD35 pKa = 3.98GDD37 pKa = 4.09DD38 pKa = 3.95SITVEE43 pKa = 3.97SEE45 pKa = 3.87EE46 pKa = 4.11FEE48 pKa = 4.28EE49 pKa = 5.04ASSEE53 pKa = 3.92EE54 pKa = 4.17LPEE57 pKa = 4.71GEE59 pKa = 4.37EE60 pKa = 3.89QGEE63 pKa = 4.28EE64 pKa = 3.95QSGEE68 pKa = 4.05EE69 pKa = 4.13SGEE72 pKa = 4.14EE73 pKa = 3.66AGEE76 pKa = 3.88AAEE79 pKa = 4.69FEE81 pKa = 4.55AVGDD85 pKa = 3.93VPEE88 pKa = 4.33EE89 pKa = 4.0LVTVTTRR96 pKa = 11.84IAEE99 pKa = 4.23SEE101 pKa = 4.49SAFNDD106 pKa = 3.47MVADD110 pKa = 4.46AAARR114 pKa = 11.84GLDD117 pKa = 3.4QEE119 pKa = 4.47SFAIISAEE127 pKa = 3.92YY128 pKa = 9.0EE129 pKa = 4.19AEE131 pKa = 4.42GEE133 pKa = 4.23LSPKK137 pKa = 10.37SYY139 pKa = 10.51EE140 pKa = 3.93ALAKK144 pKa = 10.39VGYY147 pKa = 9.13SKK149 pKa = 11.31NFIDD153 pKa = 5.67SFIAGQEE160 pKa = 4.36AIGSQYY166 pKa = 10.28MSAIQAYY173 pKa = 9.85AGGEE177 pKa = 4.02AKK179 pKa = 10.3FQAIFAHH186 pKa = 6.37LQASSPEE193 pKa = 4.08SAEE196 pKa = 3.87ALEE199 pKa = 4.14NAIGQRR205 pKa = 11.84DD206 pKa = 3.77LKK208 pKa = 10.67AVKK211 pKa = 10.48GIINLAASSVRR222 pKa = 11.84KK223 pKa = 9.76EE224 pKa = 3.89FGKK227 pKa = 9.7PAEE230 pKa = 4.13RR231 pKa = 11.84TLSKK235 pKa = 10.38RR236 pKa = 11.84ATPAKK241 pKa = 9.08VTAPKK246 pKa = 10.39AAGFEE251 pKa = 4.65SQAEE255 pKa = 4.24MVKK258 pKa = 10.68AMKK261 pKa = 10.08DD262 pKa = 3.28PKK264 pKa = 10.08YY265 pKa = 10.96GRR267 pKa = 11.84DD268 pKa = 3.13AAYY271 pKa = 8.85TRR273 pKa = 11.84SVEE276 pKa = 4.69LKK278 pKa = 10.21VLNAKK283 pKa = 9.48FF284 pKa = 3.3
Molecular weight: 30.38 kDa
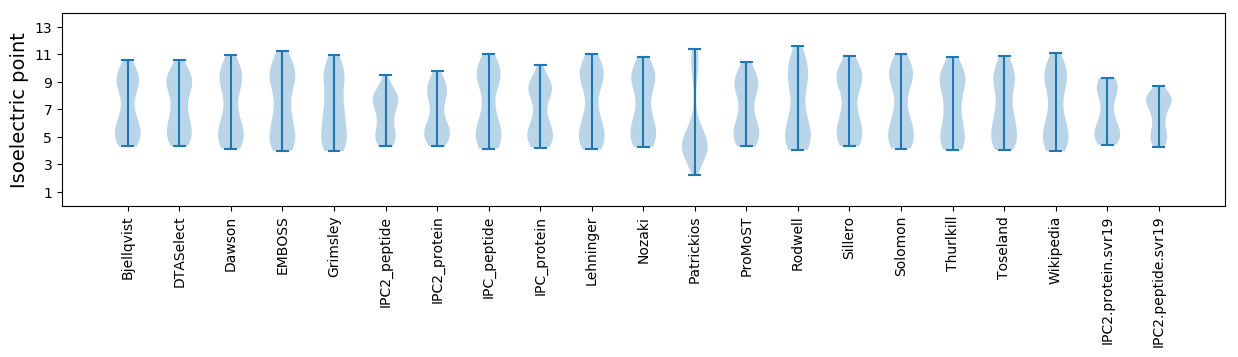
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385IFE2|A0A385IFE2_9CAUD DNA-directed RNA polymerase OS=Dickeya phage Dagda_B1 OX=2320189 PE=3 SV=1
MM1 pKa = 6.03VTAWDD6 pKa = 4.0TMSGTLRR13 pKa = 11.84LSWKK17 pKa = 9.85ALRR20 pKa = 11.84AKK22 pKa = 9.42MHH24 pKa = 5.84GRR26 pKa = 11.84PLCWRR31 pKa = 11.84NKK33 pKa = 6.06MTSKK37 pKa = 10.97SNSQNKK43 pKa = 10.01AGFIKK48 pKa = 10.0RR49 pKa = 11.84TWQRR53 pKa = 11.84TKK55 pKa = 10.99LIPLGLLMIYY65 pKa = 10.29VSLVTGCASGSQPVRR80 pKa = 11.84PLSPQVQVDD89 pKa = 3.65QALMVTPNFQEE100 pKa = 4.12TLLKK104 pKa = 10.16YY105 pKa = 10.77LSVNPDD111 pKa = 2.89VPMSGLNDD119 pKa = 3.6SKK121 pKa = 11.31RR122 pKa = 11.84PP123 pKa = 3.6
MM1 pKa = 6.03VTAWDD6 pKa = 4.0TMSGTLRR13 pKa = 11.84LSWKK17 pKa = 9.85ALRR20 pKa = 11.84AKK22 pKa = 9.42MHH24 pKa = 5.84GRR26 pKa = 11.84PLCWRR31 pKa = 11.84NKK33 pKa = 6.06MTSKK37 pKa = 10.97SNSQNKK43 pKa = 10.01AGFIKK48 pKa = 10.0RR49 pKa = 11.84TWQRR53 pKa = 11.84TKK55 pKa = 10.99LIPLGLLMIYY65 pKa = 10.29VSLVTGCASGSQPVRR80 pKa = 11.84PLSPQVQVDD89 pKa = 3.65QALMVTPNFQEE100 pKa = 4.12TLLKK104 pKa = 10.16YY105 pKa = 10.77LSVNPDD111 pKa = 2.89VPMSGLNDD119 pKa = 3.6SKK121 pKa = 11.31RR122 pKa = 11.84PP123 pKa = 3.6
Molecular weight: 13.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12766 |
30 |
1320 |
240.9 |
26.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.345 ± 0.459 | 0.995 ± 0.167 |
6.032 ± 0.213 | 6.729 ± 0.379 |
3.924 ± 0.231 | 7.418 ± 0.373 |
1.864 ± 0.192 | 5.233 ± 0.253 |
6.556 ± 0.308 | 7.966 ± 0.328 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.039 ± 0.168 | 4.692 ± 0.245 |
3.838 ± 0.183 | 4.206 ± 0.369 |
5.115 ± 0.155 | 6.431 ± 0.296 |
5.468 ± 0.272 | 6.682 ± 0.273 |
1.433 ± 0.164 | 3.031 ± 0.166 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
