
bacterium G20
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins
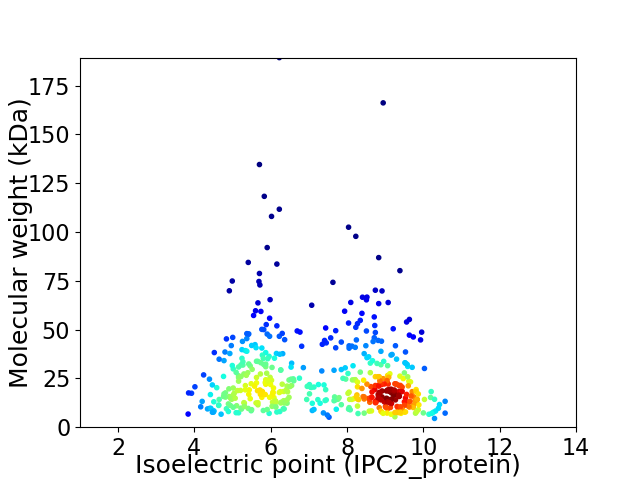
Virtual 2D-PAGE plot for 493 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
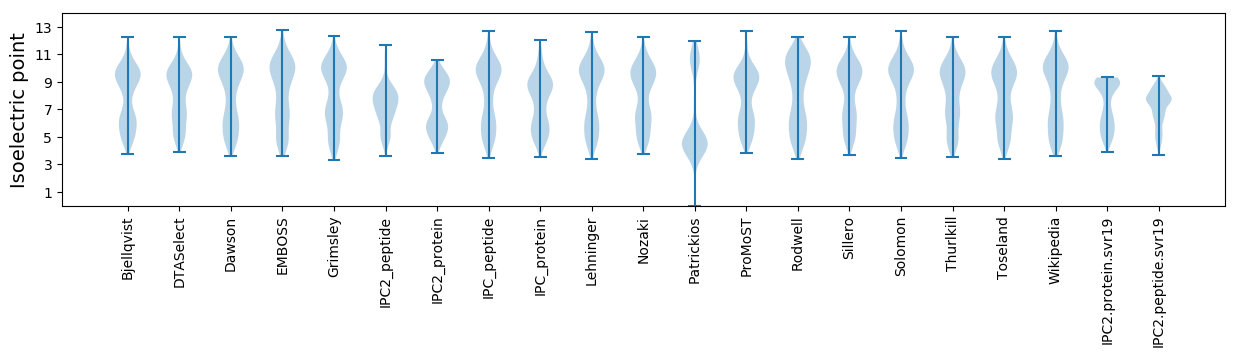
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A202DRG4|A0A202DRG4_9BACT Superoxide dismutase OS=bacterium G20 OX=1932699 GN=BVY00_02510 PE=3 SV=1
MM1 pKa = 7.77ASTFSILSSTYY12 pKa = 9.63TNTASGTTINGDD24 pKa = 3.22LGYY27 pKa = 7.57TTGPAVAPTVNGITHH42 pKa = 7.03IADD45 pKa = 3.26GTYY48 pKa = 9.91NQAGIDD54 pKa = 3.52QGTALSALNSQACTFTFPAGAVDD77 pKa = 4.5LATDD81 pKa = 4.0TSHH84 pKa = 6.57GVIGIYY90 pKa = 9.74TPGVYY95 pKa = 8.72CTTASSAASIGTAGITLSGSGTYY118 pKa = 9.8IFRR121 pKa = 11.84VNGALTTVANSAVRR135 pKa = 11.84LASSASACDD144 pKa = 3.7IFWTPTAATTLGANSTFIGTDD165 pKa = 2.6IDD167 pKa = 3.83ASGVTIGDD175 pKa = 4.32TITT178 pKa = 3.3
MM1 pKa = 7.77ASTFSILSSTYY12 pKa = 9.63TNTASGTTINGDD24 pKa = 3.22LGYY27 pKa = 7.57TTGPAVAPTVNGITHH42 pKa = 7.03IADD45 pKa = 3.26GTYY48 pKa = 9.91NQAGIDD54 pKa = 3.52QGTALSALNSQACTFTFPAGAVDD77 pKa = 4.5LATDD81 pKa = 4.0TSHH84 pKa = 6.57GVIGIYY90 pKa = 9.74TPGVYY95 pKa = 8.72CTTASSAASIGTAGITLSGSGTYY118 pKa = 9.8IFRR121 pKa = 11.84VNGALTTVANSAVRR135 pKa = 11.84LASSASACDD144 pKa = 3.7IFWTPTAATTLGANSTFIGTDD165 pKa = 2.6IDD167 pKa = 3.83ASGVTIGDD175 pKa = 4.32TITT178 pKa = 3.3
Molecular weight: 17.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A202DTP1|A0A202DTP1_9BACT Uncharacterized protein OS=bacterium G20 OX=1932699 GN=BVY00_00510 PE=4 SV=1
MM1 pKa = 7.22GQGASQPQPASEE13 pKa = 3.95TGRR16 pKa = 11.84RR17 pKa = 11.84HH18 pKa = 5.81RR19 pKa = 11.84PDD21 pKa = 3.15YY22 pKa = 11.05AILVLTALLLVIGLIVVYY40 pKa = 10.18SISPGLSASQHH51 pKa = 4.64VSQSYY56 pKa = 10.32FITKK60 pKa = 9.82QLLDD64 pKa = 3.7VALGAAAFAIAAVLPLRR81 pKa = 11.84RR82 pKa = 11.84WSAFSRR88 pKa = 11.84SLIVATVVGSVIVMITPINAIYY110 pKa = 8.51PAHH113 pKa = 6.01RR114 pKa = 11.84WIRR117 pKa = 11.84IGGFSFQVAEE127 pKa = 4.79LIKK130 pKa = 10.17LTLIVGLANFLSGQWRR146 pKa = 11.84KK147 pKa = 10.03GKK149 pKa = 10.02LADD152 pKa = 5.03FKK154 pKa = 11.23ATLKK158 pKa = 10.66PLIIVLLGVGIVVAKK173 pKa = 10.02LQSDD177 pKa = 4.27LGSAGVMIVIMAAMAFTVGIPLKK200 pKa = 10.47KK201 pKa = 10.18VALICALVTLVFALAIASSGYY222 pKa = 9.48RR223 pKa = 11.84RR224 pKa = 11.84QRR226 pKa = 11.84LATFLHH232 pKa = 6.78PSSDD236 pKa = 3.88CRR238 pKa = 11.84SSGYY242 pKa = 9.6HH243 pKa = 6.09ACQALIAVGSGGVFGEE259 pKa = 4.23GLGFGAASYY268 pKa = 10.59GYY270 pKa = 10.51LPEE273 pKa = 4.74SSNDD277 pKa = 3.58SIFAIMAQKK286 pKa = 10.44FGFIGTTLIVTIYY299 pKa = 10.56GFFITRR305 pKa = 11.84IKK307 pKa = 10.69KK308 pKa = 10.37LIEE311 pKa = 3.85RR312 pKa = 11.84TNDD315 pKa = 2.95QFSRR319 pKa = 11.84LILVGVLAWLSTQTIVNIGAMLGLLPLKK347 pKa = 10.62GITLPLISQGGTSLIFLTAALGIVFQISRR376 pKa = 11.84YY377 pKa = 6.72TSYY380 pKa = 11.53NVTEE384 pKa = 4.51PKK386 pKa = 9.88TGSEE390 pKa = 3.94QGSNTNSSSNRR401 pKa = 11.84RR402 pKa = 11.84RR403 pKa = 11.84LRR405 pKa = 11.84RR406 pKa = 11.84PYY408 pKa = 10.41NPPTISRR415 pKa = 11.84PRR417 pKa = 11.84TT418 pKa = 3.33
MM1 pKa = 7.22GQGASQPQPASEE13 pKa = 3.95TGRR16 pKa = 11.84RR17 pKa = 11.84HH18 pKa = 5.81RR19 pKa = 11.84PDD21 pKa = 3.15YY22 pKa = 11.05AILVLTALLLVIGLIVVYY40 pKa = 10.18SISPGLSASQHH51 pKa = 4.64VSQSYY56 pKa = 10.32FITKK60 pKa = 9.82QLLDD64 pKa = 3.7VALGAAAFAIAAVLPLRR81 pKa = 11.84RR82 pKa = 11.84WSAFSRR88 pKa = 11.84SLIVATVVGSVIVMITPINAIYY110 pKa = 8.51PAHH113 pKa = 6.01RR114 pKa = 11.84WIRR117 pKa = 11.84IGGFSFQVAEE127 pKa = 4.79LIKK130 pKa = 10.17LTLIVGLANFLSGQWRR146 pKa = 11.84KK147 pKa = 10.03GKK149 pKa = 10.02LADD152 pKa = 5.03FKK154 pKa = 11.23ATLKK158 pKa = 10.66PLIIVLLGVGIVVAKK173 pKa = 10.02LQSDD177 pKa = 4.27LGSAGVMIVIMAAMAFTVGIPLKK200 pKa = 10.47KK201 pKa = 10.18VALICALVTLVFALAIASSGYY222 pKa = 9.48RR223 pKa = 11.84RR224 pKa = 11.84QRR226 pKa = 11.84LATFLHH232 pKa = 6.78PSSDD236 pKa = 3.88CRR238 pKa = 11.84SSGYY242 pKa = 9.6HH243 pKa = 6.09ACQALIAVGSGGVFGEE259 pKa = 4.23GLGFGAASYY268 pKa = 10.59GYY270 pKa = 10.51LPEE273 pKa = 4.74SSNDD277 pKa = 3.58SIFAIMAQKK286 pKa = 10.44FGFIGTTLIVTIYY299 pKa = 10.56GFFITRR305 pKa = 11.84IKK307 pKa = 10.69KK308 pKa = 10.37LIEE311 pKa = 3.85RR312 pKa = 11.84TNDD315 pKa = 2.95QFSRR319 pKa = 11.84LILVGVLAWLSTQTIVNIGAMLGLLPLKK347 pKa = 10.62GITLPLISQGGTSLIFLTAALGIVFQISRR376 pKa = 11.84YY377 pKa = 6.72TSYY380 pKa = 11.53NVTEE384 pKa = 4.51PKK386 pKa = 9.88TGSEE390 pKa = 3.94QGSNTNSSSNRR401 pKa = 11.84RR402 pKa = 11.84RR403 pKa = 11.84LRR405 pKa = 11.84RR406 pKa = 11.84PYY408 pKa = 10.41NPPTISRR415 pKa = 11.84PRR417 pKa = 11.84TT418 pKa = 3.33
Molecular weight: 44.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
119114 |
38 |
1880 |
241.6 |
26.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.946 ± 0.119 | 0.541 ± 0.032 |
5.158 ± 0.104 | 5.974 ± 0.195 |
3.553 ± 0.082 | 7.224 ± 0.148 |
2.082 ± 0.058 | 6.415 ± 0.106 |
6.772 ± 0.149 | 9.705 ± 0.13 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.938 ± 0.059 | 4.096 ± 0.132 |
4.531 ± 0.11 | 4.071 ± 0.078 |
5.286 ± 0.114 | 6.531 ± 0.147 |
5.937 ± 0.21 | 7.013 ± 0.08 |
1.082 ± 0.048 | 3.146 ± 0.076 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
