
Eupatorium vein clearing virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Caulimovirus; unclassified Caulimovirus
Average proteome isoelectric point is 7.26
Get precalculated fractions of proteins
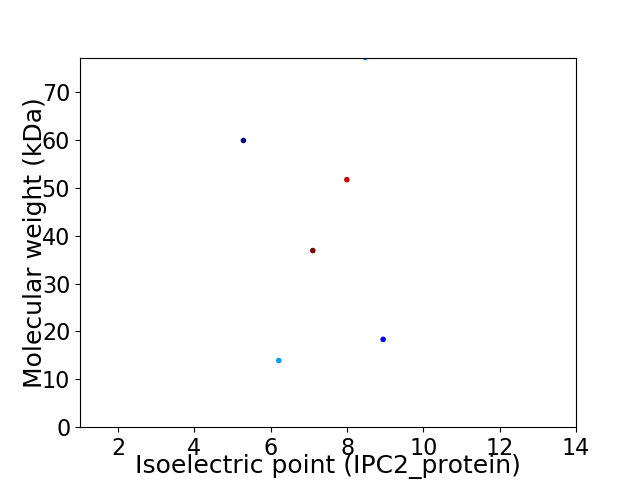
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B2D1N0|B2D1N0_9VIRU Aspartic protease OS=Eupatorium vein clearing virus OX=515444 PE=3 SV=1
MM1 pKa = 7.22LTDD4 pKa = 3.4IQKK7 pKa = 8.36VTNLFFDD14 pKa = 3.47SWKK17 pKa = 10.19EE18 pKa = 4.12GEE20 pKa = 4.02RR21 pKa = 11.84HH22 pKa = 5.07EE23 pKa = 5.41AFTSEE28 pKa = 3.73IAKK31 pKa = 10.23IMSLPEE37 pKa = 3.7EE38 pKa = 4.44EE39 pKa = 4.54ANLMLRR45 pKa = 11.84NNSHH49 pKa = 6.64LRR51 pKa = 11.84GIFQLDD57 pKa = 3.46DD58 pKa = 3.93PQNLFQLTEE67 pKa = 4.12LEE69 pKa = 4.62EE70 pKa = 4.4EE71 pKa = 4.38QSSPVYY77 pKa = 10.63DD78 pKa = 4.64SAPEE82 pKa = 3.66WDD84 pKa = 5.05EE85 pKa = 4.52YY86 pKa = 11.64DD87 pKa = 3.68DD88 pKa = 6.43DD89 pKa = 5.47YY90 pKa = 11.94ILVITNNAHH99 pKa = 7.19SDD101 pKa = 3.46DD102 pKa = 4.84GYY104 pKa = 10.26NTDD107 pKa = 3.48DD108 pKa = 4.67TINVTIPSDD117 pKa = 3.37TDD119 pKa = 3.32EE120 pKa = 4.64EE121 pKa = 4.38EE122 pKa = 4.43EE123 pKa = 4.63EE124 pKa = 4.25INLGNLRR131 pKa = 11.84PEE133 pKa = 4.25IFQDD137 pKa = 3.85DD138 pKa = 4.16EE139 pKa = 4.73ASSGVFNEE147 pKa = 4.61SGLSGGPSTSRR158 pKa = 11.84PQPRR162 pKa = 11.84PNMGDD167 pKa = 3.51PMRR170 pKa = 11.84LKK172 pKa = 10.29PDD174 pKa = 2.89TGTQVLNLDD183 pKa = 4.05CTTSFSGRR191 pKa = 11.84RR192 pKa = 11.84ALIEE196 pKa = 3.93LWKK199 pKa = 10.78KK200 pKa = 10.62EE201 pKa = 3.79MDD203 pKa = 4.26IILLTGKK210 pKa = 9.58IRR212 pKa = 11.84TAEE215 pKa = 4.04EE216 pKa = 4.59LIMLVDD222 pKa = 3.98YY223 pKa = 8.32KK224 pKa = 11.01TAGNVNAAIKK234 pKa = 9.99GYY236 pKa = 9.25SWNRR240 pKa = 11.84QFSPADD246 pKa = 3.38LLEE249 pKa = 4.24AVRR252 pKa = 11.84KK253 pKa = 9.47VLYY256 pKa = 9.76TVFLGEE262 pKa = 4.07DD263 pKa = 3.34HH264 pKa = 6.67ATQEE268 pKa = 4.27ALEE271 pKa = 4.31VAVRR275 pKa = 11.84IANAKK280 pKa = 10.5SIMTNLKK287 pKa = 9.7LCNICNVDD295 pKa = 3.49EE296 pKa = 4.83FFCTFEE302 pKa = 4.93KK303 pKa = 11.0YY304 pKa = 7.91MFRR307 pKa = 11.84IPIGEE312 pKa = 4.09HH313 pKa = 5.91PEE315 pKa = 3.73WVQMYY320 pKa = 9.61LRR322 pKa = 11.84KK323 pKa = 9.7IPFVGEE329 pKa = 3.38QAYY332 pKa = 9.76NQFMANAPEE341 pKa = 4.02TSKK344 pKa = 10.98PSLAAAHH351 pKa = 6.28RR352 pKa = 11.84VVKK355 pKa = 10.71DD356 pKa = 3.71LLNQKK361 pKa = 10.1CLDD364 pKa = 3.62ALQTKK369 pKa = 9.36KK370 pKa = 10.72LKK372 pKa = 10.67KK373 pKa = 10.36FSTKK377 pKa = 10.02CCPKK381 pKa = 10.62LIPQNLEE388 pKa = 3.6IGCPAPKK395 pKa = 10.04KK396 pKa = 6.64WTNRR400 pKa = 11.84KK401 pKa = 8.73KK402 pKa = 10.19KK403 pKa = 10.23SYY405 pKa = 10.06RR406 pKa = 11.84KK407 pKa = 9.22SSKK410 pKa = 9.16QRR412 pKa = 11.84SMTKK416 pKa = 9.7SYY418 pKa = 10.25RR419 pKa = 11.84GKK421 pKa = 10.25RR422 pKa = 11.84RR423 pKa = 11.84KK424 pKa = 6.84TTYY427 pKa = 9.63QPRR430 pKa = 11.84KK431 pKa = 8.51YY432 pKa = 10.14FRR434 pKa = 11.84KK435 pKa = 10.11KK436 pKa = 9.71SAKK439 pKa = 8.21TDD441 pKa = 3.27KK442 pKa = 10.93KK443 pKa = 10.84KK444 pKa = 10.66NCPKK448 pKa = 10.56GKK450 pKa = 9.91SSCKK454 pKa = 9.7CWICNMEE461 pKa = 3.79GHH463 pKa = 6.28YY464 pKa = 11.37ANDD467 pKa = 3.37CPEE470 pKa = 3.96RR471 pKa = 11.84NKK473 pKa = 10.59NSKK476 pKa = 7.03TVKK479 pKa = 10.15FLQTLDD485 pKa = 3.3QMGYY489 pKa = 10.41EE490 pKa = 4.04PVEE493 pKa = 4.75DD494 pKa = 3.89IFDD497 pKa = 4.13GEE499 pKa = 4.24QEE501 pKa = 4.05LFYY504 pKa = 10.67FDD506 pKa = 3.95EE507 pKa = 4.6VEE509 pKa = 4.18PGEE512 pKa = 4.2TSEE515 pKa = 4.57EE516 pKa = 4.05EE517 pKa = 4.2SSEE520 pKa = 4.54DD521 pKa = 3.52EE522 pKa = 4.14
MM1 pKa = 7.22LTDD4 pKa = 3.4IQKK7 pKa = 8.36VTNLFFDD14 pKa = 3.47SWKK17 pKa = 10.19EE18 pKa = 4.12GEE20 pKa = 4.02RR21 pKa = 11.84HH22 pKa = 5.07EE23 pKa = 5.41AFTSEE28 pKa = 3.73IAKK31 pKa = 10.23IMSLPEE37 pKa = 3.7EE38 pKa = 4.44EE39 pKa = 4.54ANLMLRR45 pKa = 11.84NNSHH49 pKa = 6.64LRR51 pKa = 11.84GIFQLDD57 pKa = 3.46DD58 pKa = 3.93PQNLFQLTEE67 pKa = 4.12LEE69 pKa = 4.62EE70 pKa = 4.4EE71 pKa = 4.38QSSPVYY77 pKa = 10.63DD78 pKa = 4.64SAPEE82 pKa = 3.66WDD84 pKa = 5.05EE85 pKa = 4.52YY86 pKa = 11.64DD87 pKa = 3.68DD88 pKa = 6.43DD89 pKa = 5.47YY90 pKa = 11.94ILVITNNAHH99 pKa = 7.19SDD101 pKa = 3.46DD102 pKa = 4.84GYY104 pKa = 10.26NTDD107 pKa = 3.48DD108 pKa = 4.67TINVTIPSDD117 pKa = 3.37TDD119 pKa = 3.32EE120 pKa = 4.64EE121 pKa = 4.38EE122 pKa = 4.43EE123 pKa = 4.63EE124 pKa = 4.25INLGNLRR131 pKa = 11.84PEE133 pKa = 4.25IFQDD137 pKa = 3.85DD138 pKa = 4.16EE139 pKa = 4.73ASSGVFNEE147 pKa = 4.61SGLSGGPSTSRR158 pKa = 11.84PQPRR162 pKa = 11.84PNMGDD167 pKa = 3.51PMRR170 pKa = 11.84LKK172 pKa = 10.29PDD174 pKa = 2.89TGTQVLNLDD183 pKa = 4.05CTTSFSGRR191 pKa = 11.84RR192 pKa = 11.84ALIEE196 pKa = 3.93LWKK199 pKa = 10.78KK200 pKa = 10.62EE201 pKa = 3.79MDD203 pKa = 4.26IILLTGKK210 pKa = 9.58IRR212 pKa = 11.84TAEE215 pKa = 4.04EE216 pKa = 4.59LIMLVDD222 pKa = 3.98YY223 pKa = 8.32KK224 pKa = 11.01TAGNVNAAIKK234 pKa = 9.99GYY236 pKa = 9.25SWNRR240 pKa = 11.84QFSPADD246 pKa = 3.38LLEE249 pKa = 4.24AVRR252 pKa = 11.84KK253 pKa = 9.47VLYY256 pKa = 9.76TVFLGEE262 pKa = 4.07DD263 pKa = 3.34HH264 pKa = 6.67ATQEE268 pKa = 4.27ALEE271 pKa = 4.31VAVRR275 pKa = 11.84IANAKK280 pKa = 10.5SIMTNLKK287 pKa = 9.7LCNICNVDD295 pKa = 3.49EE296 pKa = 4.83FFCTFEE302 pKa = 4.93KK303 pKa = 11.0YY304 pKa = 7.91MFRR307 pKa = 11.84IPIGEE312 pKa = 4.09HH313 pKa = 5.91PEE315 pKa = 3.73WVQMYY320 pKa = 9.61LRR322 pKa = 11.84KK323 pKa = 9.7IPFVGEE329 pKa = 3.38QAYY332 pKa = 9.76NQFMANAPEE341 pKa = 4.02TSKK344 pKa = 10.98PSLAAAHH351 pKa = 6.28RR352 pKa = 11.84VVKK355 pKa = 10.71DD356 pKa = 3.71LLNQKK361 pKa = 10.1CLDD364 pKa = 3.62ALQTKK369 pKa = 9.36KK370 pKa = 10.72LKK372 pKa = 10.67KK373 pKa = 10.36FSTKK377 pKa = 10.02CCPKK381 pKa = 10.62LIPQNLEE388 pKa = 3.6IGCPAPKK395 pKa = 10.04KK396 pKa = 6.64WTNRR400 pKa = 11.84KK401 pKa = 8.73KK402 pKa = 10.19KK403 pKa = 10.23SYY405 pKa = 10.06RR406 pKa = 11.84KK407 pKa = 9.22SSKK410 pKa = 9.16QRR412 pKa = 11.84SMTKK416 pKa = 9.7SYY418 pKa = 10.25RR419 pKa = 11.84GKK421 pKa = 10.25RR422 pKa = 11.84RR423 pKa = 11.84KK424 pKa = 6.84TTYY427 pKa = 9.63QPRR430 pKa = 11.84KK431 pKa = 8.51YY432 pKa = 10.14FRR434 pKa = 11.84KK435 pKa = 10.11KK436 pKa = 9.71SAKK439 pKa = 8.21TDD441 pKa = 3.27KK442 pKa = 10.93KK443 pKa = 10.84KK444 pKa = 10.66NCPKK448 pKa = 10.56GKK450 pKa = 9.91SSCKK454 pKa = 9.7CWICNMEE461 pKa = 3.79GHH463 pKa = 6.28YY464 pKa = 11.37ANDD467 pKa = 3.37CPEE470 pKa = 3.96RR471 pKa = 11.84NKK473 pKa = 10.59NSKK476 pKa = 7.03TVKK479 pKa = 10.15FLQTLDD485 pKa = 3.3QMGYY489 pKa = 10.41EE490 pKa = 4.04PVEE493 pKa = 4.75DD494 pKa = 3.89IFDD497 pKa = 4.13GEE499 pKa = 4.24QEE501 pKa = 4.05LFYY504 pKa = 10.67FDD506 pKa = 3.95EE507 pKa = 4.6VEE509 pKa = 4.18PGEE512 pKa = 4.2TSEE515 pKa = 4.57EE516 pKa = 4.05EE517 pKa = 4.2SSEE520 pKa = 4.54DD521 pKa = 3.52EE522 pKa = 4.14
Molecular weight: 59.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2D1M8|B2D1M8_9VIRU Protein 3 OS=Eupatorium vein clearing virus OX=515444 PE=3 SV=1
MM1 pKa = 7.4ANKK4 pKa = 9.31MLTYY8 pKa = 9.01PHH10 pKa = 7.47IYY12 pKa = 9.9KK13 pKa = 10.47KK14 pKa = 10.97GLIYY18 pKa = 10.2EE19 pKa = 4.74FKK21 pKa = 11.15SLDD24 pKa = 3.65DD25 pKa = 3.63KK26 pKa = 11.78SNPRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 8.37TFSSDD37 pKa = 3.1EE38 pKa = 4.0NSAGLKK44 pKa = 10.05PIIRR48 pKa = 11.84HH49 pKa = 5.87LNNINQITARR59 pKa = 11.84NWLKK63 pKa = 9.58LTKK66 pKa = 10.35LLAYY70 pKa = 10.58LGLEE74 pKa = 3.83KK75 pKa = 10.9DD76 pKa = 3.54KK77 pKa = 11.65TNGLSKK83 pKa = 10.64KK84 pKa = 9.83PSPWDD89 pKa = 3.24QLLKK93 pKa = 10.6DD94 pKa = 3.51IEE96 pKa = 4.79KK97 pKa = 10.1IFHH100 pKa = 6.82RR101 pKa = 11.84SSSSKK106 pKa = 11.22GNDD109 pKa = 3.07QTKK112 pKa = 10.38LLEE115 pKa = 4.16KK116 pKa = 10.7LEE118 pKa = 4.79EE119 pKa = 4.35ISHH122 pKa = 6.54KK123 pKa = 10.58LPKK126 pKa = 10.32AEE128 pKa = 3.82NLVTRR133 pKa = 11.84EE134 pKa = 4.07EE135 pKa = 3.89LSGIIKK141 pKa = 10.31SFHH144 pKa = 6.58EE145 pKa = 3.94EE146 pKa = 3.77LKK148 pKa = 10.38EE149 pKa = 3.94VKK151 pKa = 10.75SMIKK155 pKa = 10.23SVIGG159 pKa = 3.42
MM1 pKa = 7.4ANKK4 pKa = 9.31MLTYY8 pKa = 9.01PHH10 pKa = 7.47IYY12 pKa = 9.9KK13 pKa = 10.47KK14 pKa = 10.97GLIYY18 pKa = 10.2EE19 pKa = 4.74FKK21 pKa = 11.15SLDD24 pKa = 3.65DD25 pKa = 3.63KK26 pKa = 11.78SNPRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 8.37TFSSDD37 pKa = 3.1EE38 pKa = 4.0NSAGLKK44 pKa = 10.05PIIRR48 pKa = 11.84HH49 pKa = 5.87LNNINQITARR59 pKa = 11.84NWLKK63 pKa = 9.58LTKK66 pKa = 10.35LLAYY70 pKa = 10.58LGLEE74 pKa = 3.83KK75 pKa = 10.9DD76 pKa = 3.54KK77 pKa = 11.65TNGLSKK83 pKa = 10.64KK84 pKa = 9.83PSPWDD89 pKa = 3.24QLLKK93 pKa = 10.6DD94 pKa = 3.51IEE96 pKa = 4.79KK97 pKa = 10.1IFHH100 pKa = 6.82RR101 pKa = 11.84SSSSKK106 pKa = 11.22GNDD109 pKa = 3.07QTKK112 pKa = 10.38LLEE115 pKa = 4.16KK116 pKa = 10.7LEE118 pKa = 4.79EE119 pKa = 4.35ISHH122 pKa = 6.54KK123 pKa = 10.58LPKK126 pKa = 10.32AEE128 pKa = 3.82NLVTRR133 pKa = 11.84EE134 pKa = 4.07EE135 pKa = 3.89LSGIIKK141 pKa = 10.31SFHH144 pKa = 6.58EE145 pKa = 3.94EE146 pKa = 3.77LKK148 pKa = 10.38EE149 pKa = 3.94VKK151 pKa = 10.75SMIKK155 pKa = 10.23SVIGG159 pKa = 3.42
Molecular weight: 18.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2267 |
126 |
674 |
377.8 |
43.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.896 ± 0.628 | 1.456 ± 0.36 |
5.249 ± 0.499 | 7.322 ± 0.648 |
4.191 ± 0.418 | 4.852 ± 0.133 |
2.338 ± 0.338 | 7.543 ± 0.982 |
9.881 ± 0.685 | 8.91 ± 0.496 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.029 ± 0.274 | 6.22 ± 0.628 |
5.249 ± 0.808 | 4.102 ± 0.333 |
3.705 ± 0.43 | 7.719 ± 0.475 |
5.293 ± 0.384 | 4.808 ± 0.584 |
0.97 ± 0.219 | 3.264 ± 0.105 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
