
Aliivibrio fischeri (strain ATCC 700601 / ES114) (Vibrio fischeri)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Aliivibrio; Aliivibrio fischeri
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
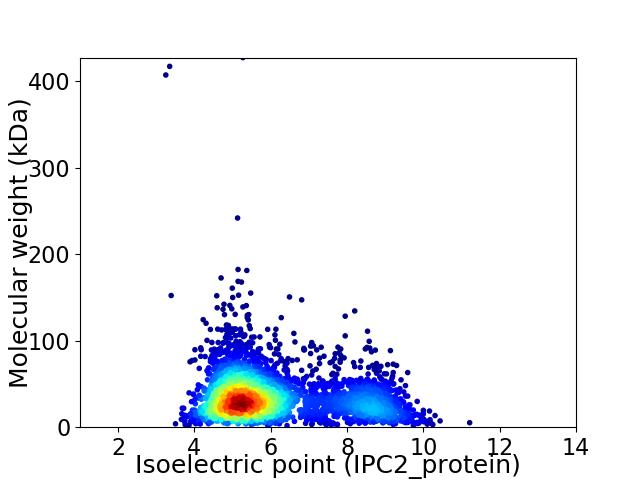
Virtual 2D-PAGE plot for 3813 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5E0B6|Q5E0B6_ALIF1 Transcription-repair-coupling factor OS=Aliivibrio fischeri (strain ATCC 700601 / ES114) OX=312309 GN=mfd PE=3 SV=2
MM1 pKa = 7.91KK2 pKa = 10.1YY3 pKa = 9.99KK4 pKa = 10.49QLIPIIALATASIPIHH20 pKa = 5.49ATIVDD25 pKa = 4.39LDD27 pKa = 3.91FSNHH31 pKa = 6.43IEE33 pKa = 4.07DD34 pKa = 4.12TNLNNSFGPSYY45 pKa = 10.23DD46 pKa = 3.37GPVMHH51 pKa = 7.04FLNVGVHH58 pKa = 5.55NGKK61 pKa = 8.48TIDD64 pKa = 3.53AKK66 pKa = 10.37ISSRR70 pKa = 11.84IIGDD74 pKa = 3.29ATFLYY79 pKa = 7.73HH80 pKa = 5.97TPNYY84 pKa = 10.08KK85 pKa = 10.25EE86 pKa = 5.4GSTQPSGDD94 pKa = 3.16IGFLYY99 pKa = 8.57QTNSPGPAGLIYY111 pKa = 10.45TFEE114 pKa = 4.47FFDD117 pKa = 4.05GTDD120 pKa = 3.54GLSGTFSIPYY130 pKa = 7.37TIPEE134 pKa = 4.21FEE136 pKa = 4.48MIGYY140 pKa = 9.73DD141 pKa = 3.27IDD143 pKa = 4.11GEE145 pKa = 4.46PVQSEE150 pKa = 3.96QVRR153 pKa = 11.84VYY155 pKa = 10.72KK156 pKa = 10.98NEE158 pKa = 3.81GFFSYY163 pKa = 8.81QTGSAGASLTAEE175 pKa = 4.28EE176 pKa = 5.0SPDD179 pKa = 3.47GLSVLFTGPGTNYY192 pKa = 10.61NEE194 pKa = 4.47TDD196 pKa = 2.7TSGAVKK202 pKa = 8.87FTYY205 pKa = 10.46KK206 pKa = 9.25NTSIVTLQFEE216 pKa = 4.8TVTASNSPLPNPIFSAFDD234 pKa = 3.9GNWDD238 pKa = 4.24LDD240 pKa = 4.07DD241 pKa = 4.0FTPPIGSSDD250 pKa = 3.44EE251 pKa = 4.46SDD253 pKa = 4.31FGDD256 pKa = 5.22APDD259 pKa = 4.5SYY261 pKa = 11.35NTLKK265 pKa = 11.3SNDD268 pKa = 3.8GAEE271 pKa = 4.25HH272 pKa = 6.88AMTSTLYY279 pKa = 10.81LGSSVDD285 pKa = 4.06ADD287 pKa = 3.51TDD289 pKa = 3.9GQPGIASNGDD299 pKa = 3.68DD300 pKa = 5.14LDD302 pKa = 4.12VDD304 pKa = 4.56GNDD307 pKa = 4.44DD308 pKa = 4.43DD309 pKa = 6.44GITILTSLEE318 pKa = 4.15KK319 pKa = 11.04GLDD322 pKa = 3.23ALINVNASGSGYY334 pKa = 9.58LQAWADD340 pKa = 2.97WDD342 pKa = 3.99MNGSFDD348 pKa = 4.34EE349 pKa = 5.0GEE351 pKa = 4.31QILTNHH357 pKa = 7.44PIVSGVQIVPIRR369 pKa = 11.84VSDD372 pKa = 3.64SATIGSVQTRR382 pKa = 11.84FRR384 pKa = 11.84LSSNPNIPSSGYY396 pKa = 9.6VGDD399 pKa = 5.05GEE401 pKa = 4.47VEE403 pKa = 4.34DD404 pKa = 4.31YY405 pKa = 11.5VFDD408 pKa = 3.77VTDD411 pKa = 4.27PGTTIQHH418 pKa = 5.47SGYY421 pKa = 8.33YY422 pKa = 8.46TAAFEE427 pKa = 5.51DD428 pKa = 3.88NWPEE432 pKa = 4.65IGDD435 pKa = 3.69FDD437 pKa = 6.1LNDD440 pKa = 3.0VVAYY444 pKa = 9.76YY445 pKa = 8.33RR446 pKa = 11.84TTIVSKK452 pKa = 10.69DD453 pKa = 3.52GEE455 pKa = 4.41VLRR458 pKa = 11.84FDD460 pKa = 3.16ITGTIMAYY468 pKa = 9.6GASYY472 pKa = 11.43GNGLGWKK479 pKa = 9.95LNGFSEE485 pKa = 4.56SDD487 pKa = 3.28INLSLARR494 pKa = 11.84LEE496 pKa = 4.47KK497 pKa = 11.05NGVTRR502 pKa = 11.84ANISPFTGEE511 pKa = 4.6DD512 pKa = 3.16KK513 pKa = 11.16SIASPGGDD521 pKa = 2.85LVVVASLNLRR531 pKa = 11.84EE532 pKa = 4.77DD533 pKa = 3.43IIINEE538 pKa = 3.98EE539 pKa = 4.36CKK541 pKa = 10.22FHH543 pKa = 6.43RR544 pKa = 11.84TNPSCSASLEE554 pKa = 4.23SEE556 pKa = 3.8QMTFSISLPFNDD568 pKa = 4.07GSEE571 pKa = 4.24PSVSSLLPLNGADD584 pKa = 3.74PFIFAAGYY592 pKa = 9.95GLYY595 pKa = 10.44HH596 pKa = 7.12GDD598 pKa = 3.88SFSTAPGTDD607 pKa = 3.9LEE609 pKa = 4.31IHH611 pKa = 6.09TADD614 pKa = 4.54FPPTSRR620 pKa = 11.84GTLVSSFYY628 pKa = 11.12GIAQDD633 pKa = 4.99DD634 pKa = 4.23SDD636 pKa = 4.44PATSKK641 pKa = 10.94YY642 pKa = 10.75YY643 pKa = 9.64RR644 pKa = 11.84TTGNLPWGILISSPWNHH661 pKa = 6.24PSEE664 pKa = 4.69YY665 pKa = 9.65IDD667 pKa = 5.02IGDD670 pKa = 4.87AYY672 pKa = 10.14PDD674 pKa = 3.58FAEE677 pKa = 4.36WATSGGTEE685 pKa = 3.62KK686 pKa = 8.31TTWYY690 pKa = 10.63LNPTASNTWSTADD703 pKa = 3.13
MM1 pKa = 7.91KK2 pKa = 10.1YY3 pKa = 9.99KK4 pKa = 10.49QLIPIIALATASIPIHH20 pKa = 5.49ATIVDD25 pKa = 4.39LDD27 pKa = 3.91FSNHH31 pKa = 6.43IEE33 pKa = 4.07DD34 pKa = 4.12TNLNNSFGPSYY45 pKa = 10.23DD46 pKa = 3.37GPVMHH51 pKa = 7.04FLNVGVHH58 pKa = 5.55NGKK61 pKa = 8.48TIDD64 pKa = 3.53AKK66 pKa = 10.37ISSRR70 pKa = 11.84IIGDD74 pKa = 3.29ATFLYY79 pKa = 7.73HH80 pKa = 5.97TPNYY84 pKa = 10.08KK85 pKa = 10.25EE86 pKa = 5.4GSTQPSGDD94 pKa = 3.16IGFLYY99 pKa = 8.57QTNSPGPAGLIYY111 pKa = 10.45TFEE114 pKa = 4.47FFDD117 pKa = 4.05GTDD120 pKa = 3.54GLSGTFSIPYY130 pKa = 7.37TIPEE134 pKa = 4.21FEE136 pKa = 4.48MIGYY140 pKa = 9.73DD141 pKa = 3.27IDD143 pKa = 4.11GEE145 pKa = 4.46PVQSEE150 pKa = 3.96QVRR153 pKa = 11.84VYY155 pKa = 10.72KK156 pKa = 10.98NEE158 pKa = 3.81GFFSYY163 pKa = 8.81QTGSAGASLTAEE175 pKa = 4.28EE176 pKa = 5.0SPDD179 pKa = 3.47GLSVLFTGPGTNYY192 pKa = 10.61NEE194 pKa = 4.47TDD196 pKa = 2.7TSGAVKK202 pKa = 8.87FTYY205 pKa = 10.46KK206 pKa = 9.25NTSIVTLQFEE216 pKa = 4.8TVTASNSPLPNPIFSAFDD234 pKa = 3.9GNWDD238 pKa = 4.24LDD240 pKa = 4.07DD241 pKa = 4.0FTPPIGSSDD250 pKa = 3.44EE251 pKa = 4.46SDD253 pKa = 4.31FGDD256 pKa = 5.22APDD259 pKa = 4.5SYY261 pKa = 11.35NTLKK265 pKa = 11.3SNDD268 pKa = 3.8GAEE271 pKa = 4.25HH272 pKa = 6.88AMTSTLYY279 pKa = 10.81LGSSVDD285 pKa = 4.06ADD287 pKa = 3.51TDD289 pKa = 3.9GQPGIASNGDD299 pKa = 3.68DD300 pKa = 5.14LDD302 pKa = 4.12VDD304 pKa = 4.56GNDD307 pKa = 4.44DD308 pKa = 4.43DD309 pKa = 6.44GITILTSLEE318 pKa = 4.15KK319 pKa = 11.04GLDD322 pKa = 3.23ALINVNASGSGYY334 pKa = 9.58LQAWADD340 pKa = 2.97WDD342 pKa = 3.99MNGSFDD348 pKa = 4.34EE349 pKa = 5.0GEE351 pKa = 4.31QILTNHH357 pKa = 7.44PIVSGVQIVPIRR369 pKa = 11.84VSDD372 pKa = 3.64SATIGSVQTRR382 pKa = 11.84FRR384 pKa = 11.84LSSNPNIPSSGYY396 pKa = 9.6VGDD399 pKa = 5.05GEE401 pKa = 4.47VEE403 pKa = 4.34DD404 pKa = 4.31YY405 pKa = 11.5VFDD408 pKa = 3.77VTDD411 pKa = 4.27PGTTIQHH418 pKa = 5.47SGYY421 pKa = 8.33YY422 pKa = 8.46TAAFEE427 pKa = 5.51DD428 pKa = 3.88NWPEE432 pKa = 4.65IGDD435 pKa = 3.69FDD437 pKa = 6.1LNDD440 pKa = 3.0VVAYY444 pKa = 9.76YY445 pKa = 8.33RR446 pKa = 11.84TTIVSKK452 pKa = 10.69DD453 pKa = 3.52GEE455 pKa = 4.41VLRR458 pKa = 11.84FDD460 pKa = 3.16ITGTIMAYY468 pKa = 9.6GASYY472 pKa = 11.43GNGLGWKK479 pKa = 9.95LNGFSEE485 pKa = 4.56SDD487 pKa = 3.28INLSLARR494 pKa = 11.84LEE496 pKa = 4.47KK497 pKa = 11.05NGVTRR502 pKa = 11.84ANISPFTGEE511 pKa = 4.6DD512 pKa = 3.16KK513 pKa = 11.16SIASPGGDD521 pKa = 2.85LVVVASLNLRR531 pKa = 11.84EE532 pKa = 4.77DD533 pKa = 3.43IIINEE538 pKa = 3.98EE539 pKa = 4.36CKK541 pKa = 10.22FHH543 pKa = 6.43RR544 pKa = 11.84TNPSCSASLEE554 pKa = 4.23SEE556 pKa = 3.8QMTFSISLPFNDD568 pKa = 4.07GSEE571 pKa = 4.24PSVSSLLPLNGADD584 pKa = 3.74PFIFAAGYY592 pKa = 9.95GLYY595 pKa = 10.44HH596 pKa = 7.12GDD598 pKa = 3.88SFSTAPGTDD607 pKa = 3.9LEE609 pKa = 4.31IHH611 pKa = 6.09TADD614 pKa = 4.54FPPTSRR620 pKa = 11.84GTLVSSFYY628 pKa = 11.12GIAQDD633 pKa = 4.99DD634 pKa = 4.23SDD636 pKa = 4.44PATSKK641 pKa = 10.94YY642 pKa = 10.75YY643 pKa = 9.64RR644 pKa = 11.84TTGNLPWGILISSPWNHH661 pKa = 6.24PSEE664 pKa = 4.69YY665 pKa = 9.65IDD667 pKa = 5.02IGDD670 pKa = 4.87AYY672 pKa = 10.14PDD674 pKa = 3.58FAEE677 pKa = 4.36WATSGGTEE685 pKa = 3.62KK686 pKa = 8.31TTWYY690 pKa = 10.63LNPTASNTWSTADD703 pKa = 3.13
Molecular weight: 75.62 kDa
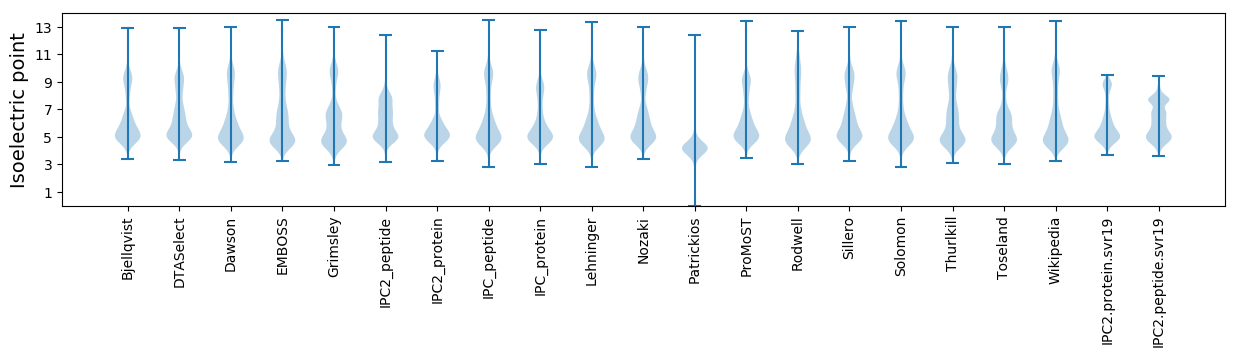
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B1WN16|B1WN16_ALIF1 Uncharacterized protein OS=Aliivibrio fischeri (strain ATCC 700601 / ES114) OX=312309 GN=VF_2608 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84NVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSKK44 pKa = 10.84
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84NVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSKK44 pKa = 10.84
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1236819 |
15 |
3971 |
324.4 |
36.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.975 ± 0.036 | 1.014 ± 0.015 |
5.486 ± 0.036 | 6.572 ± 0.04 |
4.249 ± 0.026 | 6.545 ± 0.038 |
2.164 ± 0.021 | 7.036 ± 0.035 |
5.882 ± 0.04 | 10.201 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.697 ± 0.021 | 4.741 ± 0.032 |
3.699 ± 0.025 | 4.254 ± 0.032 |
4.038 ± 0.034 | 6.823 ± 0.035 |
5.573 ± 0.039 | 6.719 ± 0.029 |
1.189 ± 0.016 | 3.145 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
