
Arrabida virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; unclassified Phlebovirus
Average proteome isoelectric point is 8.21
Get precalculated fractions of proteins
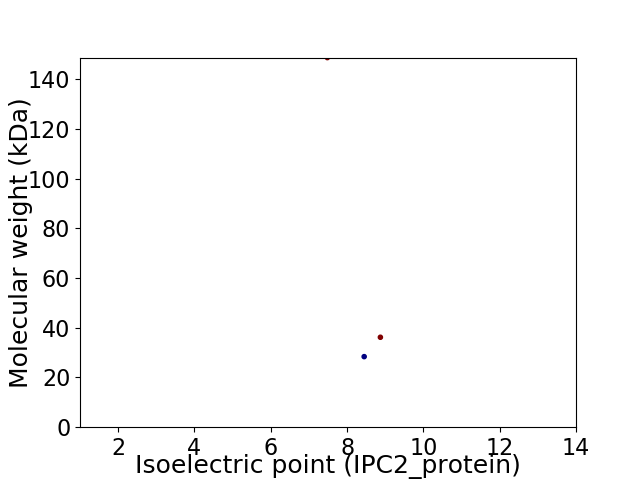
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
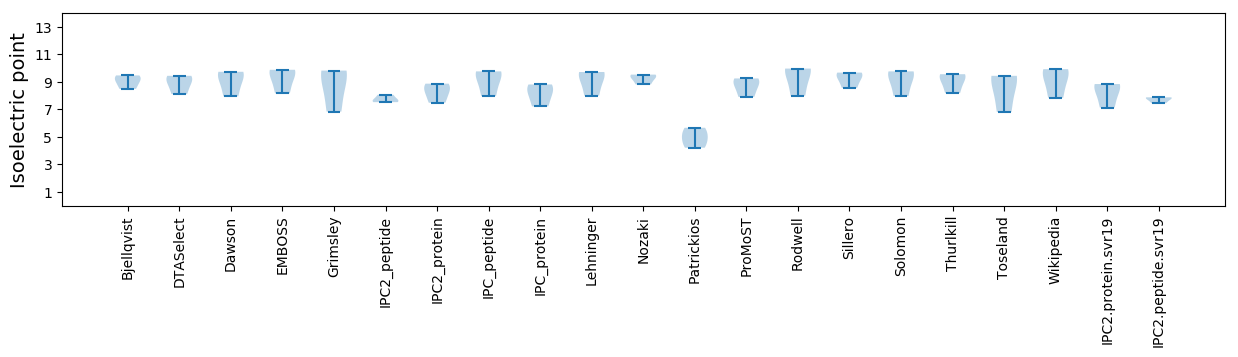
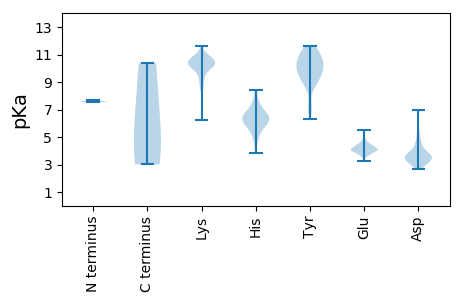
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A125QXV5|A0A125QXV5_9VIRU Nucleocapsid protein OS=Arrabida virus OX=1457322 GN=NP PE=3 SV=1
MM1 pKa = 7.54FYY3 pKa = 11.2AKK5 pKa = 10.37FILLLCFVQGALLKK19 pKa = 8.9YY20 pKa = 8.87TIISKK25 pKa = 10.39VGSSTYY31 pKa = 10.67KK32 pKa = 10.08RR33 pKa = 11.84VCLSHH38 pKa = 6.2TTDD41 pKa = 5.1LDD43 pKa = 3.68TLLDD47 pKa = 3.57MTSRR51 pKa = 11.84EE52 pKa = 3.86MSNIKK57 pKa = 8.91LTGGSCSLGDD67 pKa = 3.89LTKK70 pKa = 10.16PDD72 pKa = 4.14CSPDD76 pKa = 3.23FFSNYY81 pKa = 7.52VANIQSSNTKK91 pKa = 8.5ITGKK95 pKa = 9.69FQDD98 pKa = 3.64IIGEE102 pKa = 4.16RR103 pKa = 11.84EE104 pKa = 3.89MEE106 pKa = 4.23VTNDD110 pKa = 3.4VVPDD114 pKa = 3.66SFGPGPHH121 pKa = 7.13DD122 pKa = 4.02CVSFDD127 pKa = 5.38KK128 pKa = 11.2KK129 pKa = 10.78PFWSDD134 pKa = 3.34LLPTSVSKK142 pKa = 11.09EE143 pKa = 3.85EE144 pKa = 4.02PKK146 pKa = 10.64PIISTLSVLPAAPPIKK162 pKa = 10.33DD163 pKa = 3.66FSHH166 pKa = 6.34KK167 pKa = 10.33VVEE170 pKa = 4.18AEE172 pKa = 4.08EE173 pKa = 4.09MVKK176 pKa = 10.39EE177 pKa = 4.23IEE179 pKa = 4.21GEE181 pKa = 4.01LMKK184 pKa = 10.84EE185 pKa = 4.14KK186 pKa = 10.35EE187 pKa = 4.06RR188 pKa = 11.84NSAKK192 pKa = 10.41EE193 pKa = 3.58EE194 pKa = 3.81EE195 pKa = 4.27ARR197 pKa = 11.84QRR199 pKa = 11.84MRR201 pKa = 11.84EE202 pKa = 4.1LEE204 pKa = 4.51DD205 pKa = 4.23DD206 pKa = 5.18LRR208 pKa = 11.84KK209 pKa = 9.6QWAAEE214 pKa = 3.9RR215 pKa = 11.84MAKK218 pKa = 10.28QNMSSVLEE226 pKa = 4.06RR227 pKa = 11.84EE228 pKa = 4.18RR229 pKa = 11.84QEE231 pKa = 4.04KK232 pKa = 10.15QSLEE236 pKa = 3.95RR237 pKa = 11.84RR238 pKa = 11.84MHH240 pKa = 6.13EE241 pKa = 3.67VDD243 pKa = 2.96KK244 pKa = 10.96KK245 pKa = 11.0LRR247 pKa = 11.84ASEE250 pKa = 4.82DD251 pKa = 3.21RR252 pKa = 11.84AQKK255 pKa = 8.44YY256 pKa = 7.12QEE258 pKa = 3.97RR259 pKa = 11.84AEE261 pKa = 4.12QAEE264 pKa = 4.34KK265 pKa = 10.77SHH267 pKa = 6.52SKK269 pKa = 10.45LLSRR273 pKa = 11.84SGAATTMATVVFTSLLASGLAVVTEE298 pKa = 4.05LEE300 pKa = 4.41KK301 pKa = 10.72RR302 pKa = 11.84DD303 pKa = 3.93TNHH306 pKa = 7.15LLNRR310 pKa = 11.84PGNGVYY316 pKa = 8.88MANADD321 pKa = 3.79SLIQTEE327 pKa = 4.2CTLNYY332 pKa = 9.91GQRR335 pKa = 11.84CLAWEE340 pKa = 4.04LQITPVRR347 pKa = 11.84YY348 pKa = 8.79PFFTSNVDD356 pKa = 3.13KK357 pKa = 11.34YY358 pKa = 11.59SLLEE362 pKa = 4.72SITEE366 pKa = 4.12TPSILEE372 pKa = 4.44KK373 pKa = 11.15NNEE376 pKa = 4.06SCQLATTSGSLKK388 pKa = 9.16TCAKK392 pKa = 9.09EE393 pKa = 3.26ASTIKK398 pKa = 10.23KK399 pKa = 9.56HH400 pKa = 5.78CTDD403 pKa = 3.49DD404 pKa = 3.19VRR406 pKa = 11.84AFFFINSVGKK416 pKa = 8.52LTIVRR421 pKa = 11.84CADD424 pKa = 3.2NHH426 pKa = 6.6ILSEE430 pKa = 4.33DD431 pKa = 3.8CNFCISKK438 pKa = 10.37ARR440 pKa = 11.84NSNQNMFMPVQDD452 pKa = 4.98AFCQRR457 pKa = 11.84QGSDD461 pKa = 2.95TAPAIRR467 pKa = 11.84YY468 pKa = 9.34SKK470 pKa = 10.12DD471 pKa = 2.7ICSIGAIKK479 pKa = 10.29IKK481 pKa = 9.94QCHH484 pKa = 5.83KK485 pKa = 9.42ATSKK489 pKa = 10.16FEE491 pKa = 4.07RR492 pKa = 11.84MGFISVGQKK501 pKa = 10.14KK502 pKa = 10.43LYY504 pKa = 10.14IEE506 pKa = 4.12EE507 pKa = 4.02LKK509 pKa = 10.77LRR511 pKa = 11.84YY512 pKa = 9.03RR513 pKa = 11.84QEE515 pKa = 3.83YY516 pKa = 10.96DD517 pKa = 3.09PDD519 pKa = 3.2QFMCYY524 pKa = 9.48KK525 pKa = 9.82IKK527 pKa = 10.71SSSPLKK533 pKa = 10.06YY534 pKa = 10.31EE535 pKa = 3.88KK536 pKa = 10.79VSSKK540 pKa = 10.34KK541 pKa = 10.4CKK543 pKa = 10.16EE544 pKa = 3.56VDD546 pKa = 2.77TGGNQKK552 pKa = 10.42CSGDD556 pKa = 3.3EE557 pKa = 4.21HH558 pKa = 6.41FCNRR562 pKa = 11.84YY563 pKa = 8.97PCDD566 pKa = 3.3TANPEE571 pKa = 4.1AHH573 pKa = 6.8CLTRR577 pKa = 11.84KK578 pKa = 9.84HH579 pKa = 5.85SAIAEE584 pKa = 4.18VNIGGVWIRR593 pKa = 11.84PKK595 pKa = 10.41CVGYY599 pKa = 11.04EE600 pKa = 3.78MVLVKK605 pKa = 10.54RR606 pKa = 11.84SGLKK610 pKa = 10.27SEE612 pKa = 4.73DD613 pKa = 3.45LSIRR617 pKa = 11.84DD618 pKa = 4.18CPSCLWEE625 pKa = 4.4CKK627 pKa = 9.71KK628 pKa = 11.02SRR630 pKa = 11.84ITIKK634 pKa = 8.91THH636 pKa = 4.71GPKK639 pKa = 9.62IVSATACSHH648 pKa = 6.42GSCKK652 pKa = 10.54SVMQKK657 pKa = 10.4PSTFVEE663 pKa = 3.97IPYY666 pKa = 9.59PGNSEE671 pKa = 4.55IIGGDD676 pKa = 2.71IGVHH680 pKa = 3.87MTEE683 pKa = 4.64RR684 pKa = 11.84GSPSNIHH691 pKa = 5.98VRR693 pKa = 11.84THH695 pKa = 6.23CPPRR699 pKa = 11.84DD700 pKa = 3.54SCDD703 pKa = 3.33VSDD706 pKa = 6.22CMFCVHH712 pKa = 7.5GILNYY717 pKa = 9.83QCHH720 pKa = 5.56TLASALTIATIVSAVLVVVYY740 pKa = 9.02FLLARR745 pKa = 11.84SKK747 pKa = 10.67RR748 pKa = 11.84AVKK751 pKa = 10.28FITSLLIVPFKK762 pKa = 10.45WLTLLSIWVVKK773 pKa = 8.77NWKK776 pKa = 10.0RR777 pKa = 11.84KK778 pKa = 6.2MSKK781 pKa = 10.21LARR784 pKa = 11.84ATNNAIGWDD793 pKa = 3.44QDD795 pKa = 3.74VEE797 pKa = 4.41RR798 pKa = 11.84GRR800 pKa = 11.84PRR802 pKa = 11.84AAAGHH807 pKa = 5.2QNQVARR813 pKa = 11.84YY814 pKa = 6.34TFYY817 pKa = 11.27GATILSLMTSGLCCTEE833 pKa = 3.99SVIAEE838 pKa = 4.24SNIMQCTTSDD848 pKa = 3.23STTTCKK854 pKa = 10.76ASGTIILRR862 pKa = 11.84LGPIGSEE869 pKa = 3.66SCLILKK875 pKa = 8.76GLRR878 pKa = 11.84DD879 pKa = 3.66SEE881 pKa = 4.34KK882 pKa = 9.99QFISIRR888 pKa = 11.84TVSSEE893 pKa = 3.93LTCRR897 pKa = 11.84EE898 pKa = 3.82GDD900 pKa = 4.01SFWTSLYY907 pKa = 10.6TPICLSSRR915 pKa = 11.84RR916 pKa = 11.84CHH918 pKa = 7.36LMGEE922 pKa = 4.53CTGDD926 pKa = 4.18NCLKK930 pKa = 10.07WQDD933 pKa = 3.69NKK935 pKa = 10.62TSSEE939 pKa = 4.17FTGRR943 pKa = 11.84THH945 pKa = 8.41SEE947 pKa = 3.71VLHH950 pKa = 5.89EE951 pKa = 4.49NKK953 pKa = 10.16CFEE956 pKa = 3.93QSGGIGYY963 pKa = 9.83GCFNVNPSCLMVHH976 pKa = 6.67SYY978 pKa = 10.96LKK980 pKa = 10.27PIYY983 pKa = 10.18KK984 pKa = 10.13NAFKK988 pKa = 10.91VFRR991 pKa = 11.84CVAWNHH997 pKa = 4.89RR998 pKa = 11.84VKK1000 pKa = 10.88LAITTHH1006 pKa = 6.09KK1007 pKa = 10.65KK1008 pKa = 10.6SFDD1011 pKa = 3.55VTLMAMSTQPTDD1023 pKa = 2.68WGSIGLILDD1032 pKa = 3.72SEE1034 pKa = 5.38GITGTNSYY1042 pKa = 11.42SFMKK1046 pKa = 10.65YY1047 pKa = 10.13GVGSFAIIDD1056 pKa = 3.82EE1057 pKa = 4.58PFSIEE1062 pKa = 3.8PRR1064 pKa = 11.84KK1065 pKa = 10.54GFLGEE1070 pKa = 4.13VRR1072 pKa = 11.84CPTEE1076 pKa = 3.66EE1077 pKa = 4.14TAVKK1081 pKa = 10.43ASPSCKK1087 pKa = 9.63AAPQIIEE1094 pKa = 4.1YY1095 pKa = 9.21HH1096 pKa = 6.52PEE1098 pKa = 3.71MDD1100 pKa = 3.25TVEE1103 pKa = 5.38CITNMMDD1110 pKa = 3.78PMAIFNRR1117 pKa = 11.84GSLPQVRR1124 pKa = 11.84DD1125 pKa = 3.23GVTFAQSIEE1134 pKa = 4.3KK1135 pKa = 10.69NSVQALTTGEE1145 pKa = 3.85IKK1147 pKa = 10.77ASVRR1151 pKa = 11.84LVLDD1155 pKa = 4.4DD1156 pKa = 4.57YY1157 pKa = 10.99EE1158 pKa = 4.65VEE1160 pKa = 4.45YY1161 pKa = 10.77KK1162 pKa = 10.83VSEE1165 pKa = 4.12NDD1167 pKa = 3.62CDD1169 pKa = 3.75STFINITGCYY1179 pKa = 9.56SCDD1182 pKa = 2.99EE1183 pKa = 4.23GARR1186 pKa = 11.84LCVRR1190 pKa = 11.84IRR1192 pKa = 11.84TLGDD1196 pKa = 3.47SIYY1199 pKa = 10.62HH1200 pKa = 5.27FTDD1203 pKa = 3.23SEE1205 pKa = 4.44DD1206 pKa = 3.42MMNVMFKK1213 pKa = 10.76VNSSVTEE1220 pKa = 4.07YY1221 pKa = 10.17CTVLHH1226 pKa = 6.48FSKK1229 pKa = 10.47PVVNFEE1235 pKa = 4.03GKK1237 pKa = 9.74YY1238 pKa = 10.5DD1239 pKa = 3.69CGKK1242 pKa = 10.08SRR1244 pKa = 11.84KK1245 pKa = 9.19PMIIKK1250 pKa = 8.61GTLIAMAPHH1259 pKa = 7.86DD1260 pKa = 4.63DD1261 pKa = 3.84RR1262 pKa = 11.84VHH1264 pKa = 6.88SGGSSIVVNPKK1275 pKa = 10.39SKK1277 pKa = 10.72GVDD1280 pKa = 3.82FLGWLSGLSSWLGGPLKK1297 pKa = 10.05TVLTVLGFLILGLVFVIVVIFLVRR1321 pKa = 11.84VGIRR1325 pKa = 11.84QALLKK1330 pKa = 10.58KK1331 pKa = 10.0MKK1333 pKa = 10.4
MM1 pKa = 7.54FYY3 pKa = 11.2AKK5 pKa = 10.37FILLLCFVQGALLKK19 pKa = 8.9YY20 pKa = 8.87TIISKK25 pKa = 10.39VGSSTYY31 pKa = 10.67KK32 pKa = 10.08RR33 pKa = 11.84VCLSHH38 pKa = 6.2TTDD41 pKa = 5.1LDD43 pKa = 3.68TLLDD47 pKa = 3.57MTSRR51 pKa = 11.84EE52 pKa = 3.86MSNIKK57 pKa = 8.91LTGGSCSLGDD67 pKa = 3.89LTKK70 pKa = 10.16PDD72 pKa = 4.14CSPDD76 pKa = 3.23FFSNYY81 pKa = 7.52VANIQSSNTKK91 pKa = 8.5ITGKK95 pKa = 9.69FQDD98 pKa = 3.64IIGEE102 pKa = 4.16RR103 pKa = 11.84EE104 pKa = 3.89MEE106 pKa = 4.23VTNDD110 pKa = 3.4VVPDD114 pKa = 3.66SFGPGPHH121 pKa = 7.13DD122 pKa = 4.02CVSFDD127 pKa = 5.38KK128 pKa = 11.2KK129 pKa = 10.78PFWSDD134 pKa = 3.34LLPTSVSKK142 pKa = 11.09EE143 pKa = 3.85EE144 pKa = 4.02PKK146 pKa = 10.64PIISTLSVLPAAPPIKK162 pKa = 10.33DD163 pKa = 3.66FSHH166 pKa = 6.34KK167 pKa = 10.33VVEE170 pKa = 4.18AEE172 pKa = 4.08EE173 pKa = 4.09MVKK176 pKa = 10.39EE177 pKa = 4.23IEE179 pKa = 4.21GEE181 pKa = 4.01LMKK184 pKa = 10.84EE185 pKa = 4.14KK186 pKa = 10.35EE187 pKa = 4.06RR188 pKa = 11.84NSAKK192 pKa = 10.41EE193 pKa = 3.58EE194 pKa = 3.81EE195 pKa = 4.27ARR197 pKa = 11.84QRR199 pKa = 11.84MRR201 pKa = 11.84EE202 pKa = 4.1LEE204 pKa = 4.51DD205 pKa = 4.23DD206 pKa = 5.18LRR208 pKa = 11.84KK209 pKa = 9.6QWAAEE214 pKa = 3.9RR215 pKa = 11.84MAKK218 pKa = 10.28QNMSSVLEE226 pKa = 4.06RR227 pKa = 11.84EE228 pKa = 4.18RR229 pKa = 11.84QEE231 pKa = 4.04KK232 pKa = 10.15QSLEE236 pKa = 3.95RR237 pKa = 11.84RR238 pKa = 11.84MHH240 pKa = 6.13EE241 pKa = 3.67VDD243 pKa = 2.96KK244 pKa = 10.96KK245 pKa = 11.0LRR247 pKa = 11.84ASEE250 pKa = 4.82DD251 pKa = 3.21RR252 pKa = 11.84AQKK255 pKa = 8.44YY256 pKa = 7.12QEE258 pKa = 3.97RR259 pKa = 11.84AEE261 pKa = 4.12QAEE264 pKa = 4.34KK265 pKa = 10.77SHH267 pKa = 6.52SKK269 pKa = 10.45LLSRR273 pKa = 11.84SGAATTMATVVFTSLLASGLAVVTEE298 pKa = 4.05LEE300 pKa = 4.41KK301 pKa = 10.72RR302 pKa = 11.84DD303 pKa = 3.93TNHH306 pKa = 7.15LLNRR310 pKa = 11.84PGNGVYY316 pKa = 8.88MANADD321 pKa = 3.79SLIQTEE327 pKa = 4.2CTLNYY332 pKa = 9.91GQRR335 pKa = 11.84CLAWEE340 pKa = 4.04LQITPVRR347 pKa = 11.84YY348 pKa = 8.79PFFTSNVDD356 pKa = 3.13KK357 pKa = 11.34YY358 pKa = 11.59SLLEE362 pKa = 4.72SITEE366 pKa = 4.12TPSILEE372 pKa = 4.44KK373 pKa = 11.15NNEE376 pKa = 4.06SCQLATTSGSLKK388 pKa = 9.16TCAKK392 pKa = 9.09EE393 pKa = 3.26ASTIKK398 pKa = 10.23KK399 pKa = 9.56HH400 pKa = 5.78CTDD403 pKa = 3.49DD404 pKa = 3.19VRR406 pKa = 11.84AFFFINSVGKK416 pKa = 8.52LTIVRR421 pKa = 11.84CADD424 pKa = 3.2NHH426 pKa = 6.6ILSEE430 pKa = 4.33DD431 pKa = 3.8CNFCISKK438 pKa = 10.37ARR440 pKa = 11.84NSNQNMFMPVQDD452 pKa = 4.98AFCQRR457 pKa = 11.84QGSDD461 pKa = 2.95TAPAIRR467 pKa = 11.84YY468 pKa = 9.34SKK470 pKa = 10.12DD471 pKa = 2.7ICSIGAIKK479 pKa = 10.29IKK481 pKa = 9.94QCHH484 pKa = 5.83KK485 pKa = 9.42ATSKK489 pKa = 10.16FEE491 pKa = 4.07RR492 pKa = 11.84MGFISVGQKK501 pKa = 10.14KK502 pKa = 10.43LYY504 pKa = 10.14IEE506 pKa = 4.12EE507 pKa = 4.02LKK509 pKa = 10.77LRR511 pKa = 11.84YY512 pKa = 9.03RR513 pKa = 11.84QEE515 pKa = 3.83YY516 pKa = 10.96DD517 pKa = 3.09PDD519 pKa = 3.2QFMCYY524 pKa = 9.48KK525 pKa = 9.82IKK527 pKa = 10.71SSSPLKK533 pKa = 10.06YY534 pKa = 10.31EE535 pKa = 3.88KK536 pKa = 10.79VSSKK540 pKa = 10.34KK541 pKa = 10.4CKK543 pKa = 10.16EE544 pKa = 3.56VDD546 pKa = 2.77TGGNQKK552 pKa = 10.42CSGDD556 pKa = 3.3EE557 pKa = 4.21HH558 pKa = 6.41FCNRR562 pKa = 11.84YY563 pKa = 8.97PCDD566 pKa = 3.3TANPEE571 pKa = 4.1AHH573 pKa = 6.8CLTRR577 pKa = 11.84KK578 pKa = 9.84HH579 pKa = 5.85SAIAEE584 pKa = 4.18VNIGGVWIRR593 pKa = 11.84PKK595 pKa = 10.41CVGYY599 pKa = 11.04EE600 pKa = 3.78MVLVKK605 pKa = 10.54RR606 pKa = 11.84SGLKK610 pKa = 10.27SEE612 pKa = 4.73DD613 pKa = 3.45LSIRR617 pKa = 11.84DD618 pKa = 4.18CPSCLWEE625 pKa = 4.4CKK627 pKa = 9.71KK628 pKa = 11.02SRR630 pKa = 11.84ITIKK634 pKa = 8.91THH636 pKa = 4.71GPKK639 pKa = 9.62IVSATACSHH648 pKa = 6.42GSCKK652 pKa = 10.54SVMQKK657 pKa = 10.4PSTFVEE663 pKa = 3.97IPYY666 pKa = 9.59PGNSEE671 pKa = 4.55IIGGDD676 pKa = 2.71IGVHH680 pKa = 3.87MTEE683 pKa = 4.64RR684 pKa = 11.84GSPSNIHH691 pKa = 5.98VRR693 pKa = 11.84THH695 pKa = 6.23CPPRR699 pKa = 11.84DD700 pKa = 3.54SCDD703 pKa = 3.33VSDD706 pKa = 6.22CMFCVHH712 pKa = 7.5GILNYY717 pKa = 9.83QCHH720 pKa = 5.56TLASALTIATIVSAVLVVVYY740 pKa = 9.02FLLARR745 pKa = 11.84SKK747 pKa = 10.67RR748 pKa = 11.84AVKK751 pKa = 10.28FITSLLIVPFKK762 pKa = 10.45WLTLLSIWVVKK773 pKa = 8.77NWKK776 pKa = 10.0RR777 pKa = 11.84KK778 pKa = 6.2MSKK781 pKa = 10.21LARR784 pKa = 11.84ATNNAIGWDD793 pKa = 3.44QDD795 pKa = 3.74VEE797 pKa = 4.41RR798 pKa = 11.84GRR800 pKa = 11.84PRR802 pKa = 11.84AAAGHH807 pKa = 5.2QNQVARR813 pKa = 11.84YY814 pKa = 6.34TFYY817 pKa = 11.27GATILSLMTSGLCCTEE833 pKa = 3.99SVIAEE838 pKa = 4.24SNIMQCTTSDD848 pKa = 3.23STTTCKK854 pKa = 10.76ASGTIILRR862 pKa = 11.84LGPIGSEE869 pKa = 3.66SCLILKK875 pKa = 8.76GLRR878 pKa = 11.84DD879 pKa = 3.66SEE881 pKa = 4.34KK882 pKa = 9.99QFISIRR888 pKa = 11.84TVSSEE893 pKa = 3.93LTCRR897 pKa = 11.84EE898 pKa = 3.82GDD900 pKa = 4.01SFWTSLYY907 pKa = 10.6TPICLSSRR915 pKa = 11.84RR916 pKa = 11.84CHH918 pKa = 7.36LMGEE922 pKa = 4.53CTGDD926 pKa = 4.18NCLKK930 pKa = 10.07WQDD933 pKa = 3.69NKK935 pKa = 10.62TSSEE939 pKa = 4.17FTGRR943 pKa = 11.84THH945 pKa = 8.41SEE947 pKa = 3.71VLHH950 pKa = 5.89EE951 pKa = 4.49NKK953 pKa = 10.16CFEE956 pKa = 3.93QSGGIGYY963 pKa = 9.83GCFNVNPSCLMVHH976 pKa = 6.67SYY978 pKa = 10.96LKK980 pKa = 10.27PIYY983 pKa = 10.18KK984 pKa = 10.13NAFKK988 pKa = 10.91VFRR991 pKa = 11.84CVAWNHH997 pKa = 4.89RR998 pKa = 11.84VKK1000 pKa = 10.88LAITTHH1006 pKa = 6.09KK1007 pKa = 10.65KK1008 pKa = 10.6SFDD1011 pKa = 3.55VTLMAMSTQPTDD1023 pKa = 2.68WGSIGLILDD1032 pKa = 3.72SEE1034 pKa = 5.38GITGTNSYY1042 pKa = 11.42SFMKK1046 pKa = 10.65YY1047 pKa = 10.13GVGSFAIIDD1056 pKa = 3.82EE1057 pKa = 4.58PFSIEE1062 pKa = 3.8PRR1064 pKa = 11.84KK1065 pKa = 10.54GFLGEE1070 pKa = 4.13VRR1072 pKa = 11.84CPTEE1076 pKa = 3.66EE1077 pKa = 4.14TAVKK1081 pKa = 10.43ASPSCKK1087 pKa = 9.63AAPQIIEE1094 pKa = 4.1YY1095 pKa = 9.21HH1096 pKa = 6.52PEE1098 pKa = 3.71MDD1100 pKa = 3.25TVEE1103 pKa = 5.38CITNMMDD1110 pKa = 3.78PMAIFNRR1117 pKa = 11.84GSLPQVRR1124 pKa = 11.84DD1125 pKa = 3.23GVTFAQSIEE1134 pKa = 4.3KK1135 pKa = 10.69NSVQALTTGEE1145 pKa = 3.85IKK1147 pKa = 10.77ASVRR1151 pKa = 11.84LVLDD1155 pKa = 4.4DD1156 pKa = 4.57YY1157 pKa = 10.99EE1158 pKa = 4.65VEE1160 pKa = 4.45YY1161 pKa = 10.77KK1162 pKa = 10.83VSEE1165 pKa = 4.12NDD1167 pKa = 3.62CDD1169 pKa = 3.75STFINITGCYY1179 pKa = 9.56SCDD1182 pKa = 2.99EE1183 pKa = 4.23GARR1186 pKa = 11.84LCVRR1190 pKa = 11.84IRR1192 pKa = 11.84TLGDD1196 pKa = 3.47SIYY1199 pKa = 10.62HH1200 pKa = 5.27FTDD1203 pKa = 3.23SEE1205 pKa = 4.44DD1206 pKa = 3.42MMNVMFKK1213 pKa = 10.76VNSSVTEE1220 pKa = 4.07YY1221 pKa = 10.17CTVLHH1226 pKa = 6.48FSKK1229 pKa = 10.47PVVNFEE1235 pKa = 4.03GKK1237 pKa = 9.74YY1238 pKa = 10.5DD1239 pKa = 3.69CGKK1242 pKa = 10.08SRR1244 pKa = 11.84KK1245 pKa = 9.19PMIIKK1250 pKa = 8.61GTLIAMAPHH1259 pKa = 7.86DD1260 pKa = 4.63DD1261 pKa = 3.84RR1262 pKa = 11.84VHH1264 pKa = 6.88SGGSSIVVNPKK1275 pKa = 10.39SKK1277 pKa = 10.72GVDD1280 pKa = 3.82FLGWLSGLSSWLGGPLKK1297 pKa = 10.05TVLTVLGFLILGLVFVIVVIFLVRR1321 pKa = 11.84VGIRR1325 pKa = 11.84QALLKK1330 pKa = 10.58KK1331 pKa = 10.0MKK1333 pKa = 10.4
Molecular weight: 148.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0X9U9D7|A0A0X9U9D7_9VIRU Glycoprotein OS=Arrabida virus OX=1457322 GN=GP PE=4 SV=1
MM1 pKa = 7.66LSRR4 pKa = 11.84VVYY7 pKa = 9.9LRR9 pKa = 11.84KK10 pKa = 9.73RR11 pKa = 11.84SSDD14 pKa = 3.14RR15 pKa = 11.84KK16 pKa = 10.09KK17 pKa = 11.0GRR19 pKa = 11.84LQKK22 pKa = 10.67RR23 pKa = 11.84FFVDD27 pKa = 3.15SFLNEE32 pKa = 3.59AALFGPFSLAGGEE45 pKa = 4.49VPFTVYY51 pKa = 10.59KK52 pKa = 10.53NSVVFDD58 pKa = 3.55SRR60 pKa = 11.84PTLNHH65 pKa = 5.81YY66 pKa = 9.76LIKK69 pKa = 10.96NEE71 pKa = 4.05FPAILSGEE79 pKa = 4.38MINLPSTTLYY89 pKa = 10.54NPIMRR94 pKa = 11.84EE95 pKa = 4.07LYY97 pKa = 10.09QEE99 pKa = 4.87SIHH102 pKa = 6.47QIKK105 pKa = 10.39RR106 pKa = 11.84SNRR109 pKa = 11.84DD110 pKa = 3.3YY111 pKa = 11.17LVSALRR117 pKa = 11.84WPTGLASLEE126 pKa = 4.49FIDD129 pKa = 6.96FYY131 pKa = 11.58FEE133 pKa = 5.09DD134 pKa = 5.5FIFLSVTDD142 pKa = 4.31SYY144 pKa = 11.56ATNKK148 pKa = 9.8LLKK151 pKa = 10.51LLMKK155 pKa = 10.86ASGDD159 pKa = 3.55QSCDD163 pKa = 2.65IEE165 pKa = 4.35KK166 pKa = 10.22QIKK169 pKa = 9.42IIYY172 pKa = 9.71RR173 pKa = 11.84KK174 pKa = 9.55ILLEE178 pKa = 4.04GSKK181 pKa = 10.71FGMNCYY187 pKa = 9.91HH188 pKa = 6.64LTGADD193 pKa = 3.98IIRR196 pKa = 11.84DD197 pKa = 3.25ICIIQSARR205 pKa = 11.84VARR208 pKa = 11.84LAQKK212 pKa = 6.86TQKK215 pKa = 10.43SPSRR219 pKa = 11.84DD220 pKa = 3.52VVRR223 pKa = 11.84MIYY226 pKa = 10.24QSLSPNDD233 pKa = 4.0LLVSPKK239 pKa = 10.18HH240 pKa = 6.3SGVSTEE246 pKa = 3.97APKK249 pKa = 10.32LAEE252 pKa = 3.96MSAEE256 pKa = 3.79EE257 pKa = 4.09SFNYY261 pKa = 9.6FMAEE265 pKa = 3.78DD266 pKa = 3.8VEE268 pKa = 4.49FRR270 pKa = 11.84TMALSTFWTRR280 pKa = 11.84DD281 pKa = 2.77WPTAAEE287 pKa = 4.34TVASLRR293 pKa = 11.84QKK295 pKa = 10.0SRR297 pKa = 11.84RR298 pKa = 11.84RR299 pKa = 11.84VKK301 pKa = 10.2KK302 pKa = 9.39HH303 pKa = 5.79ASALPVSPDD312 pKa = 3.43YY313 pKa = 11.08PPLL316 pKa = 5.2
MM1 pKa = 7.66LSRR4 pKa = 11.84VVYY7 pKa = 9.9LRR9 pKa = 11.84KK10 pKa = 9.73RR11 pKa = 11.84SSDD14 pKa = 3.14RR15 pKa = 11.84KK16 pKa = 10.09KK17 pKa = 11.0GRR19 pKa = 11.84LQKK22 pKa = 10.67RR23 pKa = 11.84FFVDD27 pKa = 3.15SFLNEE32 pKa = 3.59AALFGPFSLAGGEE45 pKa = 4.49VPFTVYY51 pKa = 10.59KK52 pKa = 10.53NSVVFDD58 pKa = 3.55SRR60 pKa = 11.84PTLNHH65 pKa = 5.81YY66 pKa = 9.76LIKK69 pKa = 10.96NEE71 pKa = 4.05FPAILSGEE79 pKa = 4.38MINLPSTTLYY89 pKa = 10.54NPIMRR94 pKa = 11.84EE95 pKa = 4.07LYY97 pKa = 10.09QEE99 pKa = 4.87SIHH102 pKa = 6.47QIKK105 pKa = 10.39RR106 pKa = 11.84SNRR109 pKa = 11.84DD110 pKa = 3.3YY111 pKa = 11.17LVSALRR117 pKa = 11.84WPTGLASLEE126 pKa = 4.49FIDD129 pKa = 6.96FYY131 pKa = 11.58FEE133 pKa = 5.09DD134 pKa = 5.5FIFLSVTDD142 pKa = 4.31SYY144 pKa = 11.56ATNKK148 pKa = 9.8LLKK151 pKa = 10.51LLMKK155 pKa = 10.86ASGDD159 pKa = 3.55QSCDD163 pKa = 2.65IEE165 pKa = 4.35KK166 pKa = 10.22QIKK169 pKa = 9.42IIYY172 pKa = 9.71RR173 pKa = 11.84KK174 pKa = 9.55ILLEE178 pKa = 4.04GSKK181 pKa = 10.71FGMNCYY187 pKa = 9.91HH188 pKa = 6.64LTGADD193 pKa = 3.98IIRR196 pKa = 11.84DD197 pKa = 3.25ICIIQSARR205 pKa = 11.84VARR208 pKa = 11.84LAQKK212 pKa = 6.86TQKK215 pKa = 10.43SPSRR219 pKa = 11.84DD220 pKa = 3.52VVRR223 pKa = 11.84MIYY226 pKa = 10.24QSLSPNDD233 pKa = 4.0LLVSPKK239 pKa = 10.18HH240 pKa = 6.3SGVSTEE246 pKa = 3.97APKK249 pKa = 10.32LAEE252 pKa = 3.96MSAEE256 pKa = 3.79EE257 pKa = 4.09SFNYY261 pKa = 9.6FMAEE265 pKa = 3.78DD266 pKa = 3.8VEE268 pKa = 4.49FRR270 pKa = 11.84TMALSTFWTRR280 pKa = 11.84DD281 pKa = 2.77WPTAAEE287 pKa = 4.34TVASLRR293 pKa = 11.84QKK295 pKa = 10.0SRR297 pKa = 11.84RR298 pKa = 11.84RR299 pKa = 11.84VKK301 pKa = 10.2KK302 pKa = 9.39HH303 pKa = 5.79ASALPVSPDD312 pKa = 3.43YY313 pKa = 11.08PPLL316 pKa = 5.2
Molecular weight: 36.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1905 |
256 |
1333 |
635.0 |
71.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.247 ± 1.074 | 3.202 ± 1.269 |
4.934 ± 0.185 | 6.089 ± 0.282 |
4.199 ± 0.493 | 5.564 ± 0.695 |
1.995 ± 0.445 | 6.247 ± 0.167 |
7.559 ± 0.519 | 8.294 ± 0.738 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.202 ± 0.651 | 3.832 ± 0.24 |
4.199 ± 0.474 | 2.887 ± 0.155 |
5.302 ± 0.634 | 9.291 ± 1.072 |
6.562 ± 0.634 | 6.509 ± 0.404 |
1.102 ± 0.049 | 2.782 ± 0.433 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
