
endosymbiont of Rhynchophorus ferrugineus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; Candidatus Nardonella; Candidatus Nardonella dryophthoridicola
Average proteome isoelectric point is 8.76
Get precalculated fractions of proteins
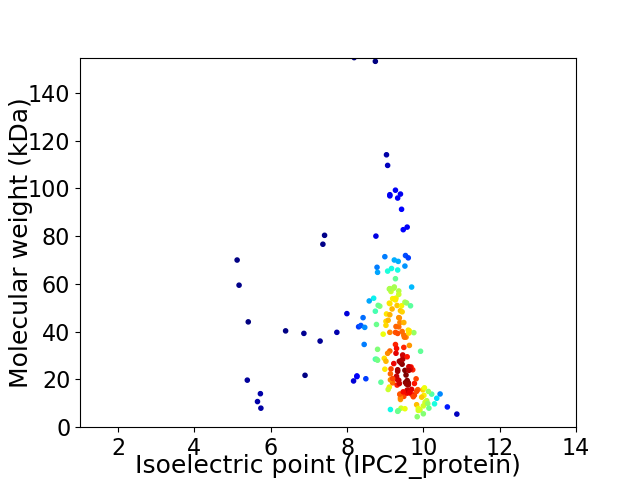
Virtual 2D-PAGE plot for 196 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
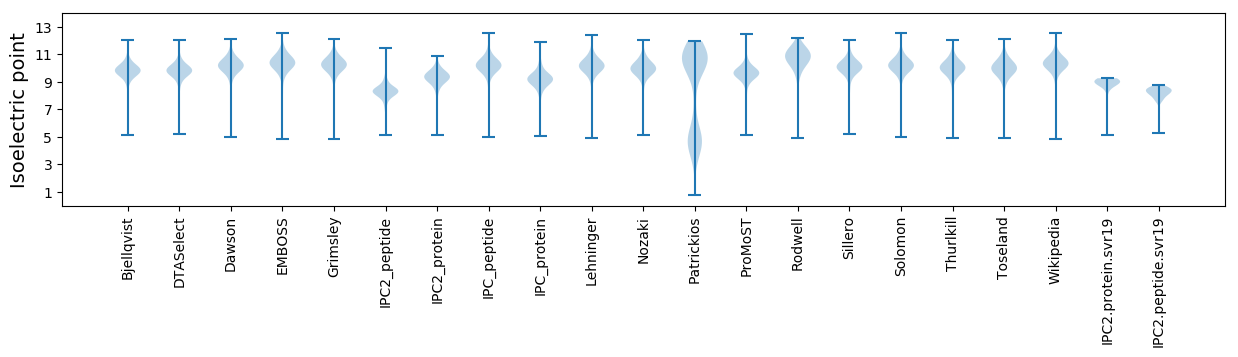
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5TGQ3|A0A2Z5TGQ3_9GAMM RNA polymerase sigma factor RpoD OS=endosymbiont of Rhynchophorus ferrugineus OX=1972133 GN=rpoD PE=3 SV=1
MM1 pKa = 7.14TSKK4 pKa = 10.27EE5 pKa = 4.11VKK7 pKa = 9.67FGNDD11 pKa = 2.87ARR13 pKa = 11.84IKK15 pKa = 9.5MLNGVNILANAVKK28 pKa = 9.39VTLGPKK34 pKa = 9.31GRR36 pKa = 11.84NVVLDD41 pKa = 3.8KK42 pKa = 11.57SFGSPLITKK51 pKa = 10.1DD52 pKa = 3.33GVSVARR58 pKa = 11.84EE59 pKa = 3.58IEE61 pKa = 4.19LEE63 pKa = 4.15DD64 pKa = 3.41KK65 pKa = 10.65FEE67 pKa = 4.45NMGAQMVKK75 pKa = 10.17EE76 pKa = 4.36VASKK80 pKa = 10.68VNDD83 pKa = 3.5MAGDD87 pKa = 3.9GTTTATLLAQSIINEE102 pKa = 4.1GLKK105 pKa = 10.74AIAAGMNPMDD115 pKa = 4.49IKK117 pKa = 10.86RR118 pKa = 11.84GIDD121 pKa = 3.12KK122 pKa = 10.73AVNYY126 pKa = 9.93AINEE130 pKa = 4.18LKK132 pKa = 10.34RR133 pKa = 11.84ISVSCSDD140 pKa = 3.91YY141 pKa = 11.27KK142 pKa = 11.27SIIQVGTISANSDD155 pKa = 3.24EE156 pKa = 5.27NIGKK160 pKa = 10.02LIADD164 pKa = 3.67AMEE167 pKa = 4.29KK168 pKa = 10.17VGKK171 pKa = 10.01EE172 pKa = 3.68GVITVEE178 pKa = 4.37EE179 pKa = 4.61GSGLNDD185 pKa = 3.08EE186 pKa = 4.78LEE188 pKa = 4.45IVEE191 pKa = 5.13GMQFDD196 pKa = 3.76RR197 pKa = 11.84GYY199 pKa = 10.76LSPYY203 pKa = 9.83FINKK207 pKa = 8.68PDD209 pKa = 3.91FNIVEE214 pKa = 4.29FDD216 pKa = 3.3NPYY219 pKa = 10.62ILLIDD224 pKa = 3.9KK225 pKa = 10.17KK226 pKa = 9.28ITGIRR231 pKa = 11.84EE232 pKa = 3.92LLPILEE238 pKa = 5.49NITKK242 pKa = 10.03SNKK245 pKa = 9.22PILIIAEE252 pKa = 4.62DD253 pKa = 3.71IEE255 pKa = 4.92GEE257 pKa = 4.08ALATLVVNNMRR268 pKa = 11.84GIFRR272 pKa = 11.84AVAVKK277 pKa = 10.65APGFGDD283 pKa = 3.21RR284 pKa = 11.84RR285 pKa = 11.84KK286 pKa = 11.2SMLQDD291 pKa = 3.1IAILTGGILISEE303 pKa = 4.69EE304 pKa = 3.71IGLDD308 pKa = 3.24ISKK311 pKa = 7.65TTLKK315 pKa = 10.97DD316 pKa = 3.03LGNARR321 pKa = 11.84KK322 pKa = 9.85IIVDD326 pKa = 3.76KK327 pKa = 11.43DD328 pKa = 3.1NTTIIDD334 pKa = 4.02GYY336 pKa = 10.82GDD338 pKa = 3.63KK339 pKa = 10.93KK340 pKa = 10.99CINQRR345 pKa = 11.84INQINKK351 pKa = 9.46EE352 pKa = 3.96KK353 pKa = 10.38EE354 pKa = 4.17QTDD357 pKa = 3.71SEE359 pKa = 4.34YY360 pKa = 11.24DD361 pKa = 3.37KK362 pKa = 11.5EE363 pKa = 4.12KK364 pKa = 10.11LQEE367 pKa = 4.59RR368 pKa = 11.84IAKK371 pKa = 10.18LSGGVALIRR380 pKa = 11.84VGAATEE386 pKa = 4.09VEE388 pKa = 4.27MKK390 pKa = 9.93EE391 pKa = 3.98KK392 pKa = 10.45KK393 pKa = 10.29SRR395 pKa = 11.84VEE397 pKa = 3.85DD398 pKa = 3.56ALNATRR404 pKa = 11.84AAVEE408 pKa = 4.16EE409 pKa = 4.64GVVAGGGVALVRR421 pKa = 11.84VSKK424 pKa = 11.01NLLNLKK430 pKa = 9.8GDD432 pKa = 3.98NEE434 pKa = 4.23DD435 pKa = 3.3QNVGIKK441 pKa = 10.09VAIKK445 pKa = 10.69SMEE448 pKa = 4.05SPLRR452 pKa = 11.84QIVSNAGEE460 pKa = 4.19EE461 pKa = 3.94PSIILNKK468 pKa = 10.1IKK470 pKa = 10.79LSDD473 pKa = 3.59GNFGYY478 pKa = 10.88NAFTEE483 pKa = 4.95EE484 pKa = 4.31YY485 pKa = 10.8GDD487 pKa = 4.04MIKK490 pKa = 9.78MGILDD495 pKa = 3.78PTKK498 pKa = 8.92VTRR501 pKa = 11.84SALQYY506 pKa = 8.67AASVAGLMITTEE518 pKa = 4.08CMITEE523 pKa = 4.32IKK525 pKa = 10.53KK526 pKa = 10.8EE527 pKa = 3.71EE528 pKa = 4.42DD529 pKa = 3.82KK530 pKa = 11.69NFDD533 pKa = 3.67TQNPNMNNMNNIMM546 pKa = 4.24
MM1 pKa = 7.14TSKK4 pKa = 10.27EE5 pKa = 4.11VKK7 pKa = 9.67FGNDD11 pKa = 2.87ARR13 pKa = 11.84IKK15 pKa = 9.5MLNGVNILANAVKK28 pKa = 9.39VTLGPKK34 pKa = 9.31GRR36 pKa = 11.84NVVLDD41 pKa = 3.8KK42 pKa = 11.57SFGSPLITKK51 pKa = 10.1DD52 pKa = 3.33GVSVARR58 pKa = 11.84EE59 pKa = 3.58IEE61 pKa = 4.19LEE63 pKa = 4.15DD64 pKa = 3.41KK65 pKa = 10.65FEE67 pKa = 4.45NMGAQMVKK75 pKa = 10.17EE76 pKa = 4.36VASKK80 pKa = 10.68VNDD83 pKa = 3.5MAGDD87 pKa = 3.9GTTTATLLAQSIINEE102 pKa = 4.1GLKK105 pKa = 10.74AIAAGMNPMDD115 pKa = 4.49IKK117 pKa = 10.86RR118 pKa = 11.84GIDD121 pKa = 3.12KK122 pKa = 10.73AVNYY126 pKa = 9.93AINEE130 pKa = 4.18LKK132 pKa = 10.34RR133 pKa = 11.84ISVSCSDD140 pKa = 3.91YY141 pKa = 11.27KK142 pKa = 11.27SIIQVGTISANSDD155 pKa = 3.24EE156 pKa = 5.27NIGKK160 pKa = 10.02LIADD164 pKa = 3.67AMEE167 pKa = 4.29KK168 pKa = 10.17VGKK171 pKa = 10.01EE172 pKa = 3.68GVITVEE178 pKa = 4.37EE179 pKa = 4.61GSGLNDD185 pKa = 3.08EE186 pKa = 4.78LEE188 pKa = 4.45IVEE191 pKa = 5.13GMQFDD196 pKa = 3.76RR197 pKa = 11.84GYY199 pKa = 10.76LSPYY203 pKa = 9.83FINKK207 pKa = 8.68PDD209 pKa = 3.91FNIVEE214 pKa = 4.29FDD216 pKa = 3.3NPYY219 pKa = 10.62ILLIDD224 pKa = 3.9KK225 pKa = 10.17KK226 pKa = 9.28ITGIRR231 pKa = 11.84EE232 pKa = 3.92LLPILEE238 pKa = 5.49NITKK242 pKa = 10.03SNKK245 pKa = 9.22PILIIAEE252 pKa = 4.62DD253 pKa = 3.71IEE255 pKa = 4.92GEE257 pKa = 4.08ALATLVVNNMRR268 pKa = 11.84GIFRR272 pKa = 11.84AVAVKK277 pKa = 10.65APGFGDD283 pKa = 3.21RR284 pKa = 11.84RR285 pKa = 11.84KK286 pKa = 11.2SMLQDD291 pKa = 3.1IAILTGGILISEE303 pKa = 4.69EE304 pKa = 3.71IGLDD308 pKa = 3.24ISKK311 pKa = 7.65TTLKK315 pKa = 10.97DD316 pKa = 3.03LGNARR321 pKa = 11.84KK322 pKa = 9.85IIVDD326 pKa = 3.76KK327 pKa = 11.43DD328 pKa = 3.1NTTIIDD334 pKa = 4.02GYY336 pKa = 10.82GDD338 pKa = 3.63KK339 pKa = 10.93KK340 pKa = 10.99CINQRR345 pKa = 11.84INQINKK351 pKa = 9.46EE352 pKa = 3.96KK353 pKa = 10.38EE354 pKa = 4.17QTDD357 pKa = 3.71SEE359 pKa = 4.34YY360 pKa = 11.24DD361 pKa = 3.37KK362 pKa = 11.5EE363 pKa = 4.12KK364 pKa = 10.11LQEE367 pKa = 4.59RR368 pKa = 11.84IAKK371 pKa = 10.18LSGGVALIRR380 pKa = 11.84VGAATEE386 pKa = 4.09VEE388 pKa = 4.27MKK390 pKa = 9.93EE391 pKa = 3.98KK392 pKa = 10.45KK393 pKa = 10.29SRR395 pKa = 11.84VEE397 pKa = 3.85DD398 pKa = 3.56ALNATRR404 pKa = 11.84AAVEE408 pKa = 4.16EE409 pKa = 4.64GVVAGGGVALVRR421 pKa = 11.84VSKK424 pKa = 11.01NLLNLKK430 pKa = 9.8GDD432 pKa = 3.98NEE434 pKa = 4.23DD435 pKa = 3.3QNVGIKK441 pKa = 10.09VAIKK445 pKa = 10.69SMEE448 pKa = 4.05SPLRR452 pKa = 11.84QIVSNAGEE460 pKa = 4.19EE461 pKa = 3.94PSIILNKK468 pKa = 10.1IKK470 pKa = 10.79LSDD473 pKa = 3.59GNFGYY478 pKa = 10.88NAFTEE483 pKa = 4.95EE484 pKa = 4.31YY485 pKa = 10.8GDD487 pKa = 4.04MIKK490 pKa = 9.78MGILDD495 pKa = 3.78PTKK498 pKa = 8.92VTRR501 pKa = 11.84SALQYY506 pKa = 8.67AASVAGLMITTEE518 pKa = 4.08CMITEE523 pKa = 4.32IKK525 pKa = 10.53KK526 pKa = 10.8EE527 pKa = 3.71EE528 pKa = 4.42DD529 pKa = 3.82KK530 pKa = 11.69NFDD533 pKa = 3.67TQNPNMNNMNNIMM546 pKa = 4.24
Molecular weight: 59.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5T9C2|A0A2Z5T9C2_9GAMM UDP-N-acetylenolpyruvoylglucosamine reductase OS=endosymbiont of Rhynchophorus ferrugineus OX=1972133 GN=murB PE=3 SV=1
MM1 pKa = 7.57SRR3 pKa = 11.84GHH5 pKa = 6.04YY6 pKa = 10.48KK7 pKa = 9.6MLFKK11 pKa = 10.75NLCNSLIKK19 pKa = 10.42YY20 pKa = 7.98EE21 pKa = 4.45KK22 pKa = 10.09IKK24 pKa = 8.54TTIKK28 pKa = 9.77RR29 pKa = 11.84AKK31 pKa = 8.86EE32 pKa = 3.44IKK34 pKa = 10.25KK35 pKa = 10.08IIEE38 pKa = 4.41PIINRR43 pKa = 11.84SKK45 pKa = 11.55VNILHH50 pKa = 6.58NKK52 pKa = 8.9RR53 pKa = 11.84IVYY56 pKa = 10.21SYY58 pKa = 11.34LNNRR62 pKa = 11.84NNVYY66 pKa = 10.93KK67 pKa = 10.73LFNDD71 pKa = 2.87IGYY74 pKa = 9.94RR75 pKa = 11.84YY76 pKa = 9.96KK77 pKa = 10.87NRR79 pKa = 11.84NGGYY83 pKa = 7.48TRR85 pKa = 11.84INKK88 pKa = 9.81LYY90 pKa = 9.2YY91 pKa = 10.15RR92 pKa = 11.84KK93 pKa = 9.95GDD95 pKa = 3.2SSLIVCISLVV105 pKa = 3.26
MM1 pKa = 7.57SRR3 pKa = 11.84GHH5 pKa = 6.04YY6 pKa = 10.48KK7 pKa = 9.6MLFKK11 pKa = 10.75NLCNSLIKK19 pKa = 10.42YY20 pKa = 7.98EE21 pKa = 4.45KK22 pKa = 10.09IKK24 pKa = 8.54TTIKK28 pKa = 9.77RR29 pKa = 11.84AKK31 pKa = 8.86EE32 pKa = 3.44IKK34 pKa = 10.25KK35 pKa = 10.08IIEE38 pKa = 4.41PIINRR43 pKa = 11.84SKK45 pKa = 11.55VNILHH50 pKa = 6.58NKK52 pKa = 8.9RR53 pKa = 11.84IVYY56 pKa = 10.21SYY58 pKa = 11.34LNNRR62 pKa = 11.84NNVYY66 pKa = 10.93KK67 pKa = 10.73LFNDD71 pKa = 2.87IGYY74 pKa = 9.94RR75 pKa = 11.84YY76 pKa = 9.96KK77 pKa = 10.87NRR79 pKa = 11.84NGGYY83 pKa = 7.48TRR85 pKa = 11.84INKK88 pKa = 9.81LYY90 pKa = 9.2YY91 pKa = 10.15RR92 pKa = 11.84KK93 pKa = 9.95GDD95 pKa = 3.2SSLIVCISLVV105 pKa = 3.26
Molecular weight: 12.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
62158 |
37 |
1338 |
317.1 |
37.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.369 ± 0.115 | 1.03 ± 0.047 |
3.94 ± 0.122 | 4.056 ± 0.144 |
5.64 ± 0.189 | 3.758 ± 0.182 |
0.981 ± 0.043 | 17.687 ± 0.278 |
13.987 ± 0.271 | 8.736 ± 0.15 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.356 ± 0.06 | 14.499 ± 0.341 |
1.815 ± 0.081 | 1.031 ± 0.066 |
2.251 ± 0.104 | 5.731 ± 0.146 |
2.714 ± 0.116 | 2.775 ± 0.13 |
0.463 ± 0.049 | 6.181 ± 0.187 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
