
Rhodospirillales bacterium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; unclassified Rhodospirillales
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
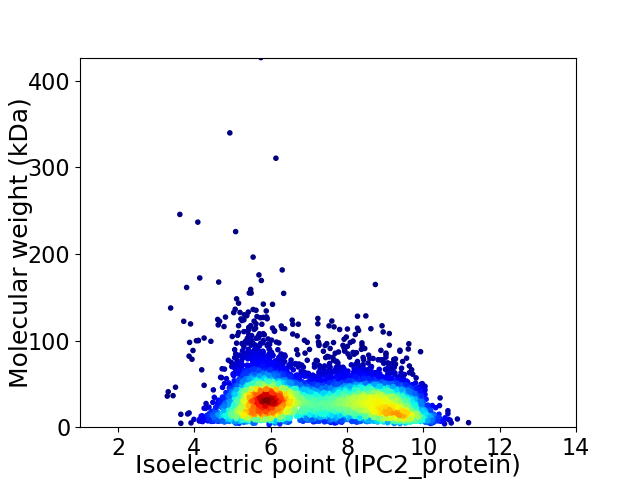
Virtual 2D-PAGE plot for 7065 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6A7MGS6|A0A6A7MGS6_9PROT DNA polymerase III subunit gamma/tau OS=Rhodospirillales bacterium OX=2026786 GN=dnaX PE=3 SV=1
MM1 pKa = 7.33PQRR4 pKa = 11.84TSRR7 pKa = 11.84SSALTRR13 pKa = 11.84STSSQTISCCDD24 pKa = 3.62LLHH27 pKa = 6.58TEE29 pKa = 4.54TRR31 pKa = 11.84QDD33 pKa = 3.48DD34 pKa = 3.54HH35 pKa = 7.73GKK37 pKa = 10.01ALNVSVSRR45 pKa = 11.84KK46 pKa = 9.15DD47 pKa = 3.11AAMPIIKK54 pKa = 10.1FGTFGNDD61 pKa = 3.21TLVGTADD68 pKa = 3.73SDD70 pKa = 3.52WLYY73 pKa = 11.64GLGGDD78 pKa = 4.89DD79 pKa = 3.77VLSGGGGEE87 pKa = 5.08DD88 pKa = 4.01YY89 pKa = 11.28LSGGDD94 pKa = 4.74DD95 pKa = 3.49NDD97 pKa = 3.36VLYY100 pKa = 11.09GGDD103 pKa = 3.56EE104 pKa = 4.27DD105 pKa = 4.58PRR107 pKa = 11.84HH108 pKa = 6.04GDD110 pKa = 3.57EE111 pKa = 5.64LDD113 pKa = 4.25GGDD116 pKa = 3.99GDD118 pKa = 4.46DD119 pKa = 3.99WLFGGAGGDD128 pKa = 3.7HH129 pKa = 7.01LLGGAGNDD137 pKa = 3.65HH138 pKa = 7.27LYY140 pKa = 11.2GGADD144 pKa = 3.36NDD146 pKa = 4.18YY147 pKa = 11.13LDD149 pKa = 4.69GGAGADD155 pKa = 3.63VMDD158 pKa = 4.87GGEE161 pKa = 3.94GLDD164 pKa = 3.08RR165 pKa = 11.84VEE167 pKa = 5.02YY168 pKa = 9.35IHH170 pKa = 6.91SPAAVTVNLTTGTGSGGDD188 pKa = 3.47AAGDD192 pKa = 3.64TLTDD196 pKa = 3.37IEE198 pKa = 4.99DD199 pKa = 3.87VIGSDD204 pKa = 4.81FADD207 pKa = 3.47TLIGNGSSNRR217 pKa = 11.84LTGGSGDD224 pKa = 4.68DD225 pKa = 3.76YY226 pKa = 11.54LSGSGGDD233 pKa = 4.07DD234 pKa = 3.58FLHH237 pKa = 6.69GGQGADD243 pKa = 3.6HH244 pKa = 6.58LHH246 pKa = 6.54GGSGSDD252 pKa = 3.14TVSYY256 pKa = 9.43TGSAAGVTVNLEE268 pKa = 4.16ANVVSGGDD276 pKa = 3.5AEE278 pKa = 5.38GDD280 pKa = 3.49TLVSIEE286 pKa = 4.28NVVGSTGADD295 pKa = 3.33VITGNAEE302 pKa = 3.86ANILSGSSGDD312 pKa = 3.75DD313 pKa = 3.09RR314 pKa = 11.84LNGGAGDD321 pKa = 3.81DD322 pKa = 4.28TLNGGQGQDD331 pKa = 3.94DD332 pKa = 5.33LIGGDD337 pKa = 3.67GADD340 pKa = 2.77TFVFSEE346 pKa = 3.89HH347 pKa = 6.3WYY349 pKa = 9.42YY350 pKa = 11.25SPPPVSDD357 pKa = 3.61TSVEE361 pKa = 4.01APDD364 pKa = 4.32YY365 pKa = 11.01IWDD368 pKa = 3.94FNHH371 pKa = 6.95AQGDD375 pKa = 4.18QIDD378 pKa = 4.5LADD381 pKa = 4.64LDD383 pKa = 4.45AVSGVPGDD391 pKa = 3.56QAFSFINTADD401 pKa = 3.66FTGSEE406 pKa = 4.23GEE408 pKa = 3.87LRR410 pKa = 11.84YY411 pKa = 9.85EE412 pKa = 4.05QFTGPFGITWTAVSGDD428 pKa = 3.5MDD430 pKa = 4.24GDD432 pKa = 3.89AVADD436 pKa = 3.98FAIVCTDD443 pKa = 4.02SINFVASDD451 pKa = 3.61FLLL454 pKa = 4.64
MM1 pKa = 7.33PQRR4 pKa = 11.84TSRR7 pKa = 11.84SSALTRR13 pKa = 11.84STSSQTISCCDD24 pKa = 3.62LLHH27 pKa = 6.58TEE29 pKa = 4.54TRR31 pKa = 11.84QDD33 pKa = 3.48DD34 pKa = 3.54HH35 pKa = 7.73GKK37 pKa = 10.01ALNVSVSRR45 pKa = 11.84KK46 pKa = 9.15DD47 pKa = 3.11AAMPIIKK54 pKa = 10.1FGTFGNDD61 pKa = 3.21TLVGTADD68 pKa = 3.73SDD70 pKa = 3.52WLYY73 pKa = 11.64GLGGDD78 pKa = 4.89DD79 pKa = 3.77VLSGGGGEE87 pKa = 5.08DD88 pKa = 4.01YY89 pKa = 11.28LSGGDD94 pKa = 4.74DD95 pKa = 3.49NDD97 pKa = 3.36VLYY100 pKa = 11.09GGDD103 pKa = 3.56EE104 pKa = 4.27DD105 pKa = 4.58PRR107 pKa = 11.84HH108 pKa = 6.04GDD110 pKa = 3.57EE111 pKa = 5.64LDD113 pKa = 4.25GGDD116 pKa = 3.99GDD118 pKa = 4.46DD119 pKa = 3.99WLFGGAGGDD128 pKa = 3.7HH129 pKa = 7.01LLGGAGNDD137 pKa = 3.65HH138 pKa = 7.27LYY140 pKa = 11.2GGADD144 pKa = 3.36NDD146 pKa = 4.18YY147 pKa = 11.13LDD149 pKa = 4.69GGAGADD155 pKa = 3.63VMDD158 pKa = 4.87GGEE161 pKa = 3.94GLDD164 pKa = 3.08RR165 pKa = 11.84VEE167 pKa = 5.02YY168 pKa = 9.35IHH170 pKa = 6.91SPAAVTVNLTTGTGSGGDD188 pKa = 3.47AAGDD192 pKa = 3.64TLTDD196 pKa = 3.37IEE198 pKa = 4.99DD199 pKa = 3.87VIGSDD204 pKa = 4.81FADD207 pKa = 3.47TLIGNGSSNRR217 pKa = 11.84LTGGSGDD224 pKa = 4.68DD225 pKa = 3.76YY226 pKa = 11.54LSGSGGDD233 pKa = 4.07DD234 pKa = 3.58FLHH237 pKa = 6.69GGQGADD243 pKa = 3.6HH244 pKa = 6.58LHH246 pKa = 6.54GGSGSDD252 pKa = 3.14TVSYY256 pKa = 9.43TGSAAGVTVNLEE268 pKa = 4.16ANVVSGGDD276 pKa = 3.5AEE278 pKa = 5.38GDD280 pKa = 3.49TLVSIEE286 pKa = 4.28NVVGSTGADD295 pKa = 3.33VITGNAEE302 pKa = 3.86ANILSGSSGDD312 pKa = 3.75DD313 pKa = 3.09RR314 pKa = 11.84LNGGAGDD321 pKa = 3.81DD322 pKa = 4.28TLNGGQGQDD331 pKa = 3.94DD332 pKa = 5.33LIGGDD337 pKa = 3.67GADD340 pKa = 2.77TFVFSEE346 pKa = 3.89HH347 pKa = 6.3WYY349 pKa = 9.42YY350 pKa = 11.25SPPPVSDD357 pKa = 3.61TSVEE361 pKa = 4.01APDD364 pKa = 4.32YY365 pKa = 11.01IWDD368 pKa = 3.94FNHH371 pKa = 6.95AQGDD375 pKa = 4.18QIDD378 pKa = 4.5LADD381 pKa = 4.64LDD383 pKa = 4.45AVSGVPGDD391 pKa = 3.56QAFSFINTADD401 pKa = 3.66FTGSEE406 pKa = 4.23GEE408 pKa = 3.87LRR410 pKa = 11.84YY411 pKa = 9.85EE412 pKa = 4.05QFTGPFGITWTAVSGDD428 pKa = 3.5MDD430 pKa = 4.24GDD432 pKa = 3.89AVADD436 pKa = 3.98FAIVCTDD443 pKa = 4.02SINFVASDD451 pKa = 3.61FLLL454 pKa = 4.64
Molecular weight: 46.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6A7LWW9|A0A6A7LWW9_9PROT Glutathione S-transferase family protein OS=Rhodospirillales bacterium OX=2026786 GN=GEV13_00565 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.33VIANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SGRR39 pKa = 11.84KK40 pKa = 8.96RR41 pKa = 11.84LSAA44 pKa = 4.09
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.33VIANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SGRR39 pKa = 11.84KK40 pKa = 8.96RR41 pKa = 11.84LSAA44 pKa = 4.09
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2124564 |
29 |
3930 |
300.7 |
32.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.536 ± 0.039 | 0.926 ± 0.009 |
5.513 ± 0.023 | 5.287 ± 0.026 |
3.654 ± 0.018 | 8.748 ± 0.034 |
2.118 ± 0.014 | 4.755 ± 0.021 |
3.386 ± 0.03 | 10.062 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.482 ± 0.015 | 2.49 ± 0.018 |
5.534 ± 0.02 | 3.061 ± 0.017 |
7.628 ± 0.037 | 5.11 ± 0.021 |
5.208 ± 0.024 | 7.692 ± 0.025 |
1.541 ± 0.012 | 2.268 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
