
Anaerotignum neopropionicum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Anaerotignum
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
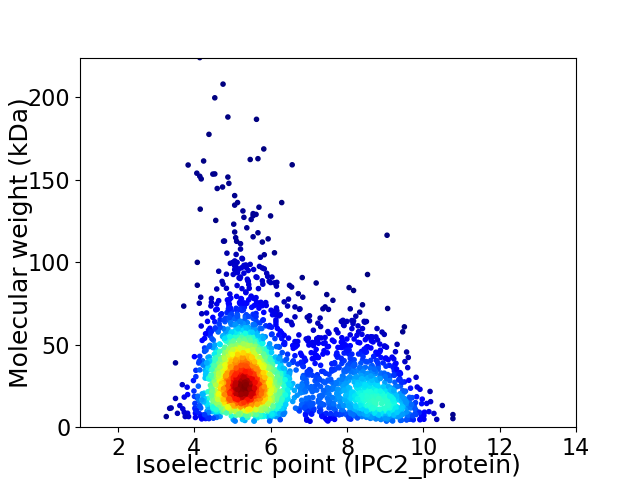
Virtual 2D-PAGE plot for 2804 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A136WEV4|A0A136WEV4_9FIRM High-affinity zinc uptake system ATP-binding protein ZnuC OS=Anaerotignum neopropionicum OX=36847 GN=znuC PE=4 SV=1
MM1 pKa = 7.23KK2 pKa = 10.6QMLALILTALLLCGMLAPSVFAEE25 pKa = 4.5VPPADD30 pKa = 3.98TEE32 pKa = 4.53TNDD35 pKa = 3.42TVMDD39 pKa = 3.82SHH41 pKa = 8.64SDD43 pKa = 3.16ADD45 pKa = 3.73TDD47 pKa = 5.09DD48 pKa = 5.32NIDD51 pKa = 3.99TITPSDD57 pKa = 3.55TGTPTDD63 pKa = 3.79TEE65 pKa = 4.44EE66 pKa = 5.53PPDD69 pKa = 3.65TDD71 pKa = 4.83EE72 pKa = 5.99NDD74 pKa = 4.08TPDD77 pKa = 3.56EE78 pKa = 4.43TQDD81 pKa = 3.14SSLPQDD87 pKa = 3.75FDD89 pKa = 3.8YY90 pKa = 10.93TDD92 pKa = 4.42SIISYY97 pKa = 7.33TIPDD101 pKa = 3.75AFTDD105 pKa = 3.87TFEE108 pKa = 4.57MDD110 pKa = 4.07CEE112 pKa = 4.3DD113 pKa = 3.38TSFTQLHH120 pKa = 6.0NALDD124 pKa = 3.98SVGISGTAKK133 pKa = 10.46KK134 pKa = 10.5DD135 pKa = 3.56DD136 pKa = 4.05TSSSLALSVIWDD148 pKa = 4.46FSTVDD153 pKa = 3.18AEE155 pKa = 4.82TPGSYY160 pKa = 10.18SAVGCISIPHH170 pKa = 6.1GAILADD176 pKa = 3.85GLEE179 pKa = 4.4STLSISVQVVAPMLAMSPGAITLTSFDD206 pKa = 3.4EE207 pKa = 4.74PYY209 pKa = 9.6RR210 pKa = 11.84TDD212 pKa = 3.2AVAFAVNTPQEE223 pKa = 4.33VLDD226 pKa = 4.3SWFADD231 pKa = 3.77CVAGFSGYY239 pKa = 10.53DD240 pKa = 3.27ADD242 pKa = 3.98GNYY245 pKa = 10.26YY246 pKa = 10.81DD247 pKa = 5.02LVSGAWSLDD256 pKa = 3.3AVDD259 pKa = 3.8TSTVGVYY266 pKa = 9.45YY267 pKa = 11.01VSTTPNLGTEE277 pKa = 4.08YY278 pKa = 10.21TLAEE282 pKa = 4.57GVSLPRR288 pKa = 11.84QLCAVSIQTPGEE300 pKa = 3.81PDD302 pKa = 3.25INCCVAGRR310 pKa = 11.84GFLHH314 pKa = 6.32FPWVISAVQQEE325 pKa = 4.25QLDD328 pKa = 4.36DD329 pKa = 3.74FTVWLRR335 pKa = 11.84QDD337 pKa = 3.51DD338 pKa = 4.83GEE340 pKa = 4.24WSSLSDD346 pKa = 3.45GFLLVSDD353 pKa = 5.07GLQLSQWIFISGSTYY368 pKa = 8.52EE369 pKa = 4.82LKK371 pKa = 9.43VTYY374 pKa = 9.81PGGQTGVLSFQFDD387 pKa = 4.13GEE389 pKa = 4.21LSIIDD394 pKa = 3.78YY395 pKa = 11.09SGGDD399 pKa = 3.41RR400 pKa = 11.84DD401 pKa = 4.62GGDD404 pKa = 3.58TNGSDD409 pKa = 3.68TGSNTQPAPTTPHH422 pKa = 6.35TPNNPQNKK430 pKa = 9.02GSHH433 pKa = 5.72NSVKK437 pKa = 9.22EE438 pKa = 4.07TLVPPPVPEE447 pKa = 4.08EE448 pKa = 3.85ATTQIPVPPTQSTPEE463 pKa = 3.85YY464 pKa = 8.58SQAAFSHH471 pKa = 5.32EE472 pKa = 4.27TEE474 pKa = 4.26GTVEE478 pKa = 4.07GTEE481 pKa = 3.9TCMAFQQPGTALTVMNPVPNKK502 pKa = 9.94IDD504 pKa = 3.62TQQEE508 pKa = 4.48TVFQSQEE515 pKa = 3.56PASPVFEE522 pKa = 4.88SYY524 pKa = 11.29SPTQTEE530 pKa = 3.82ISAMRR535 pKa = 11.84LQDD538 pKa = 3.59LCADD542 pKa = 3.96GEE544 pKa = 4.95SVVFGSGNLTVSIPSSLLLALNLSDD569 pKa = 5.56SDD571 pKa = 4.14TLTVTLTQPKK581 pKa = 10.27SNEE584 pKa = 3.42ILLAVAASGKK594 pKa = 10.5SVMEE598 pKa = 3.89LAGTVLRR605 pKa = 11.84LRR607 pKa = 11.84YY608 pKa = 8.81MPLWEE613 pKa = 4.22NSEE616 pKa = 3.79ITVGNEE622 pKa = 3.29AGEE625 pKa = 4.62KK626 pKa = 8.77ITDD629 pKa = 3.16ISYY632 pKa = 11.29DD633 pKa = 3.82DD634 pKa = 4.39EE635 pKa = 4.76LLCFAADD642 pKa = 3.55MPGTYY647 pKa = 9.64TISEE651 pKa = 4.29LANAEE656 pKa = 4.17KK657 pKa = 9.09THH659 pKa = 6.47KK660 pKa = 10.32SVSPLLPASMGLMLAGGGIAYY681 pKa = 9.06FRR683 pKa = 11.84RR684 pKa = 11.84KK685 pKa = 9.18HH686 pKa = 5.89HH687 pKa = 6.02GG688 pKa = 3.18
MM1 pKa = 7.23KK2 pKa = 10.6QMLALILTALLLCGMLAPSVFAEE25 pKa = 4.5VPPADD30 pKa = 3.98TEE32 pKa = 4.53TNDD35 pKa = 3.42TVMDD39 pKa = 3.82SHH41 pKa = 8.64SDD43 pKa = 3.16ADD45 pKa = 3.73TDD47 pKa = 5.09DD48 pKa = 5.32NIDD51 pKa = 3.99TITPSDD57 pKa = 3.55TGTPTDD63 pKa = 3.79TEE65 pKa = 4.44EE66 pKa = 5.53PPDD69 pKa = 3.65TDD71 pKa = 4.83EE72 pKa = 5.99NDD74 pKa = 4.08TPDD77 pKa = 3.56EE78 pKa = 4.43TQDD81 pKa = 3.14SSLPQDD87 pKa = 3.75FDD89 pKa = 3.8YY90 pKa = 10.93TDD92 pKa = 4.42SIISYY97 pKa = 7.33TIPDD101 pKa = 3.75AFTDD105 pKa = 3.87TFEE108 pKa = 4.57MDD110 pKa = 4.07CEE112 pKa = 4.3DD113 pKa = 3.38TSFTQLHH120 pKa = 6.0NALDD124 pKa = 3.98SVGISGTAKK133 pKa = 10.46KK134 pKa = 10.5DD135 pKa = 3.56DD136 pKa = 4.05TSSSLALSVIWDD148 pKa = 4.46FSTVDD153 pKa = 3.18AEE155 pKa = 4.82TPGSYY160 pKa = 10.18SAVGCISIPHH170 pKa = 6.1GAILADD176 pKa = 3.85GLEE179 pKa = 4.4STLSISVQVVAPMLAMSPGAITLTSFDD206 pKa = 3.4EE207 pKa = 4.74PYY209 pKa = 9.6RR210 pKa = 11.84TDD212 pKa = 3.2AVAFAVNTPQEE223 pKa = 4.33VLDD226 pKa = 4.3SWFADD231 pKa = 3.77CVAGFSGYY239 pKa = 10.53DD240 pKa = 3.27ADD242 pKa = 3.98GNYY245 pKa = 10.26YY246 pKa = 10.81DD247 pKa = 5.02LVSGAWSLDD256 pKa = 3.3AVDD259 pKa = 3.8TSTVGVYY266 pKa = 9.45YY267 pKa = 11.01VSTTPNLGTEE277 pKa = 4.08YY278 pKa = 10.21TLAEE282 pKa = 4.57GVSLPRR288 pKa = 11.84QLCAVSIQTPGEE300 pKa = 3.81PDD302 pKa = 3.25INCCVAGRR310 pKa = 11.84GFLHH314 pKa = 6.32FPWVISAVQQEE325 pKa = 4.25QLDD328 pKa = 4.36DD329 pKa = 3.74FTVWLRR335 pKa = 11.84QDD337 pKa = 3.51DD338 pKa = 4.83GEE340 pKa = 4.24WSSLSDD346 pKa = 3.45GFLLVSDD353 pKa = 5.07GLQLSQWIFISGSTYY368 pKa = 8.52EE369 pKa = 4.82LKK371 pKa = 9.43VTYY374 pKa = 9.81PGGQTGVLSFQFDD387 pKa = 4.13GEE389 pKa = 4.21LSIIDD394 pKa = 3.78YY395 pKa = 11.09SGGDD399 pKa = 3.41RR400 pKa = 11.84DD401 pKa = 4.62GGDD404 pKa = 3.58TNGSDD409 pKa = 3.68TGSNTQPAPTTPHH422 pKa = 6.35TPNNPQNKK430 pKa = 9.02GSHH433 pKa = 5.72NSVKK437 pKa = 9.22EE438 pKa = 4.07TLVPPPVPEE447 pKa = 4.08EE448 pKa = 3.85ATTQIPVPPTQSTPEE463 pKa = 3.85YY464 pKa = 8.58SQAAFSHH471 pKa = 5.32EE472 pKa = 4.27TEE474 pKa = 4.26GTVEE478 pKa = 4.07GTEE481 pKa = 3.9TCMAFQQPGTALTVMNPVPNKK502 pKa = 9.94IDD504 pKa = 3.62TQQEE508 pKa = 4.48TVFQSQEE515 pKa = 3.56PASPVFEE522 pKa = 4.88SYY524 pKa = 11.29SPTQTEE530 pKa = 3.82ISAMRR535 pKa = 11.84LQDD538 pKa = 3.59LCADD542 pKa = 3.96GEE544 pKa = 4.95SVVFGSGNLTVSIPSSLLLALNLSDD569 pKa = 5.56SDD571 pKa = 4.14TLTVTLTQPKK581 pKa = 10.27SNEE584 pKa = 3.42ILLAVAASGKK594 pKa = 10.5SVMEE598 pKa = 3.89LAGTVLRR605 pKa = 11.84LRR607 pKa = 11.84YY608 pKa = 8.81MPLWEE613 pKa = 4.22NSEE616 pKa = 3.79ITVGNEE622 pKa = 3.29AGEE625 pKa = 4.62KK626 pKa = 8.77ITDD629 pKa = 3.16ISYY632 pKa = 11.29DD633 pKa = 3.82DD634 pKa = 4.39EE635 pKa = 4.76LLCFAADD642 pKa = 3.55MPGTYY647 pKa = 9.64TISEE651 pKa = 4.29LANAEE656 pKa = 4.17KK657 pKa = 9.09THH659 pKa = 6.47KK660 pKa = 10.32SVSPLLPASMGLMLAGGGIAYY681 pKa = 9.06FRR683 pKa = 11.84RR684 pKa = 11.84KK685 pKa = 9.18HH686 pKa = 5.89HH687 pKa = 6.02GG688 pKa = 3.18
Molecular weight: 73.43 kDa
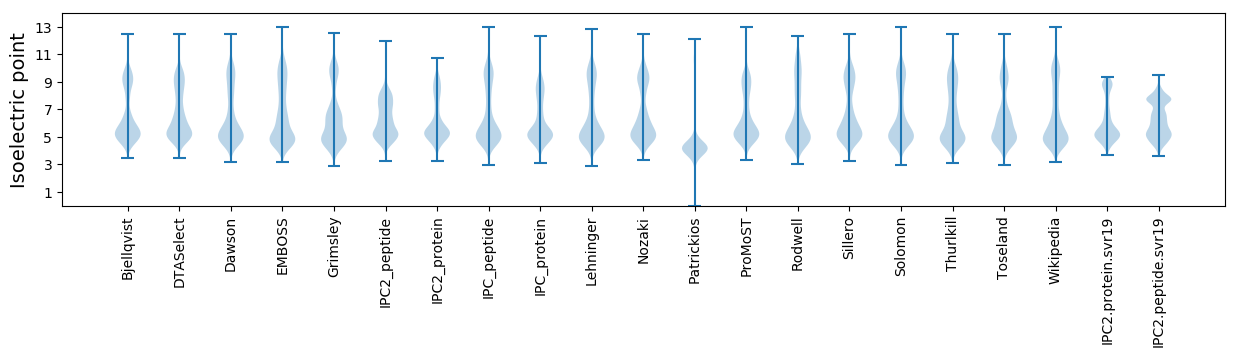
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A136WHZ4|A0A136WHZ4_9FIRM Uncharacterized protein OS=Anaerotignum neopropionicum OX=36847 GN=CLNEO_01590 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.93KK9 pKa = 7.63KK10 pKa = 9.62QRR12 pKa = 11.84SRR14 pKa = 11.84EE15 pKa = 3.67HH16 pKa = 6.32GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MLTSNGRR28 pKa = 11.84KK29 pKa = 9.11VLAARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.23KK37 pKa = 9.87GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.93KK9 pKa = 7.63KK10 pKa = 9.62QRR12 pKa = 11.84SRR14 pKa = 11.84EE15 pKa = 3.67HH16 pKa = 6.32GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MLTSNGRR28 pKa = 11.84KK29 pKa = 9.11VLAARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.23KK37 pKa = 9.87GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
838418 |
30 |
2151 |
299.0 |
33.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.562 ± 0.052 | 1.404 ± 0.019 |
5.214 ± 0.036 | 7.478 ± 0.054 |
4.464 ± 0.04 | 7.227 ± 0.05 |
1.626 ± 0.019 | 7.814 ± 0.045 |
7.1 ± 0.04 | 9.202 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.073 ± 0.023 | 4.585 ± 0.031 |
3.263 ± 0.029 | 3.312 ± 0.027 |
3.921 ± 0.035 | 5.786 ± 0.04 |
5.546 ± 0.055 | 6.856 ± 0.036 |
0.848 ± 0.016 | 3.719 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
